-
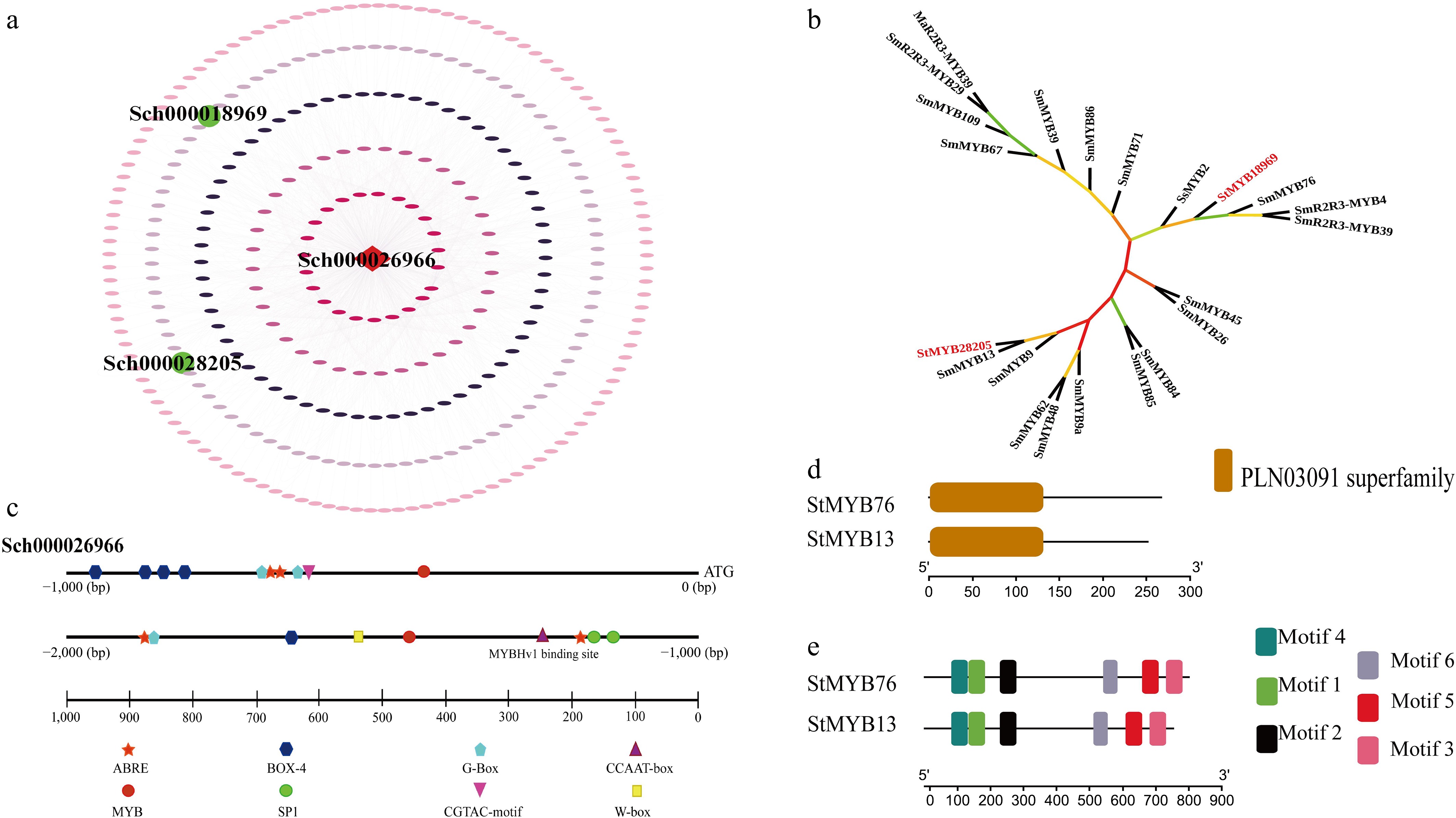
Figure 1.
Screening and identification of StMYB transcription factors. (a) The regulatory network schematic illustrates the functional core of StLS-interacting transcription factors. The regulatory network was established utilizing 1,192 binding sites identified by the FIMO tool, with green nodes denoting StMYB transcription factors and square nodes signifying the target gene StLS. (b) Phylogenetic analysis of StMYBs. The maximum likelihood method (bootstrap = 1,000) demonstrated that StMYB76/StMYB13 constitutes a distinct evolutionary branch in conjunction with Salvia miltiorrhiza SmMYB76/StMYB13, thus affirming the conserved terpenoid regulatory function within the Labiatae family. Evolutionary distances suggest that both display a strong affinity for MYBs linked to mint monoterpene synthesis (branch length < 0.2). (c) Distribution map of cis-elements in the StLS promoter. Three categories of functional modules were identified within 2,000 bp of the promoter region: metabolic regulatory elements (MYB-Hv1, G-box), stress-responsive elements (ABRE, MBS), and cross-species conserved motifs exhibiting greater than 75% homology to barley MYB-Hv1. (d), (e) Structural domain and motif composition of StMYBs. Analysis of conserved structural domains indicated that both possess a characteristic R2R3-MYB structural domain at the N-terminus (PLN03091 superfamily, E-value < 1 × 10−15). Six conserved motifs were identified using MEME, with motif 1 (N-terminal α-helix), and motif 4 (C-terminal transcriptional activation domain) forming the functional core.
-
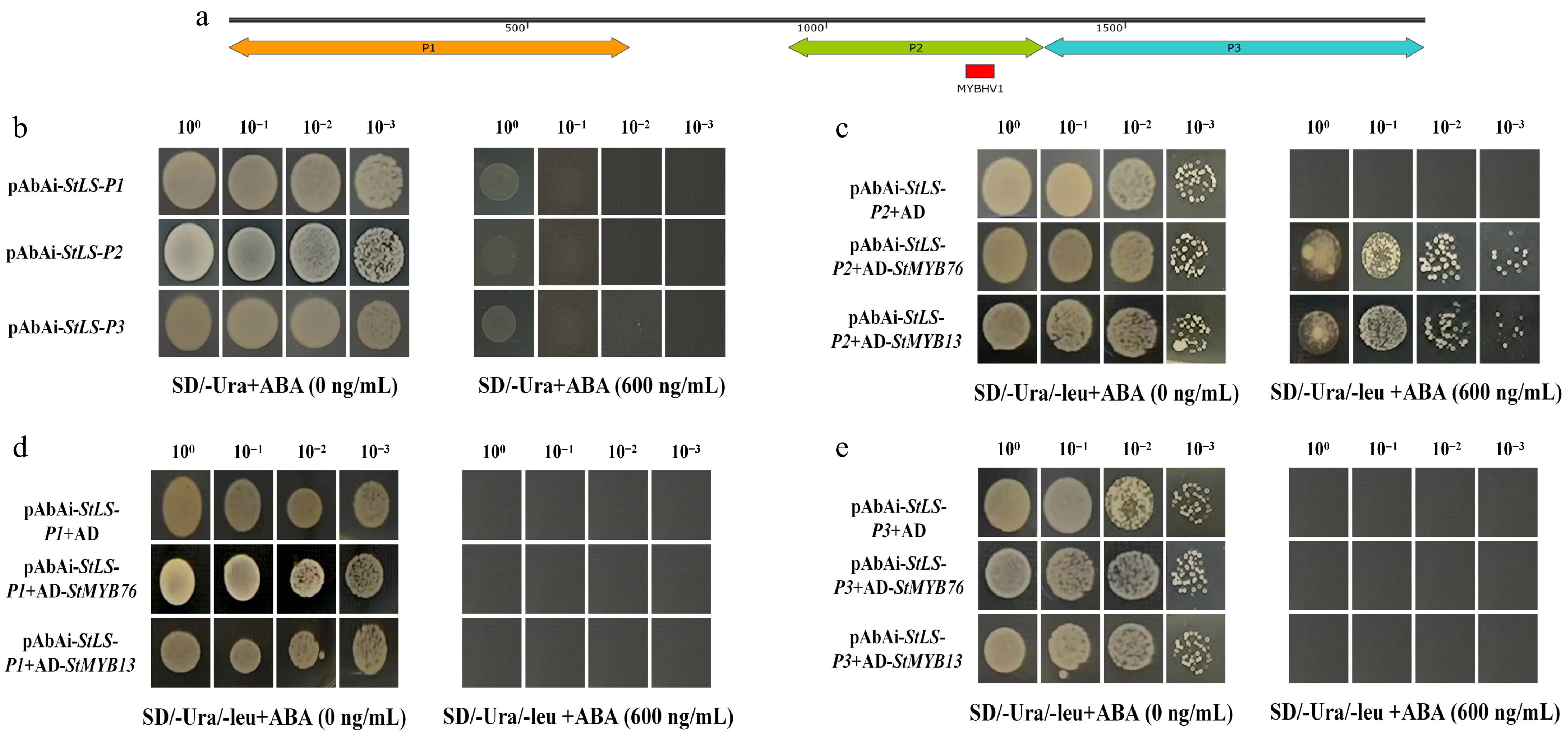
Figure 2.
Using a yeast one-hybrid system, transcription factor-promoter interactions are validated. (a) Diagrammatic representation of the StLS gene's shortened 2,000 bp promoter region upstream of the ATG start site, which includes the P1–P3 functional segments. (b) The self-activating effect of the decoy vector pAbAi-StLS-P1/P2/P3 was detected; at a concentration of 600 ng/mL AbA, no non-specific activation was seen. (c)–(e) StMYB transcription factors' binding to various promoter segments are characterized. StMYB76 and StMYB13 specifically activate the reporter system that contains the P2 fragment, while the negative control shows no growth signal.
-
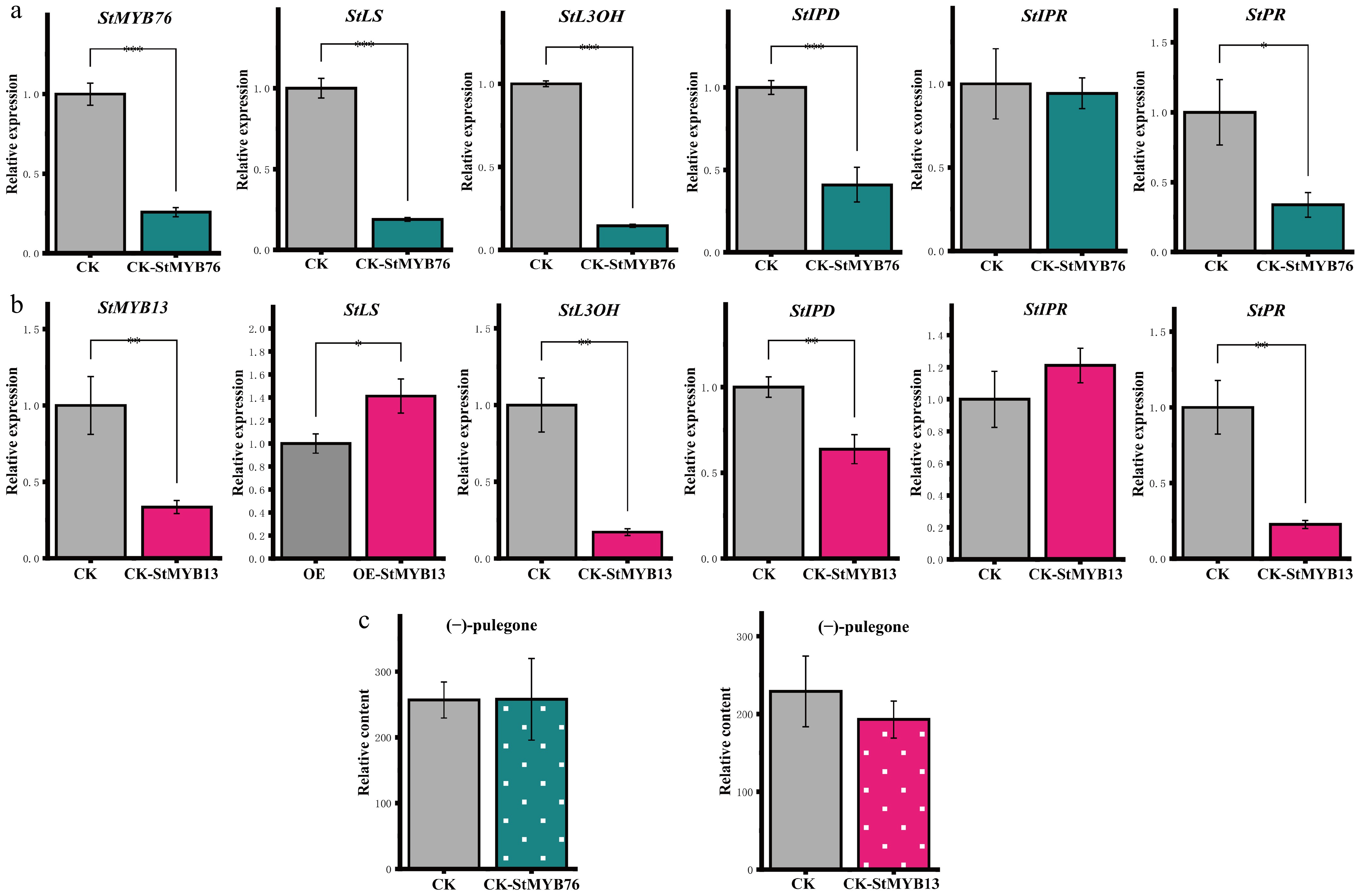
Figure 3.
Impact of StMYBs gene silencing on the (−)-pulegone biosynthetic pathway. (a), (b) The silencing of the StMYB76/StMYB13 gene leads to changes in the expression levels of critical genes in the (−)-pulegone biosynthesis pathway. (c) Changes in the concentration of (−)-pulegone monoterpene in S. tenuifolia following the silencing of StMYBs. * p < 0.05, ** p < 0.01, *** p < 0.001, 'ns' denotes not significant.
-
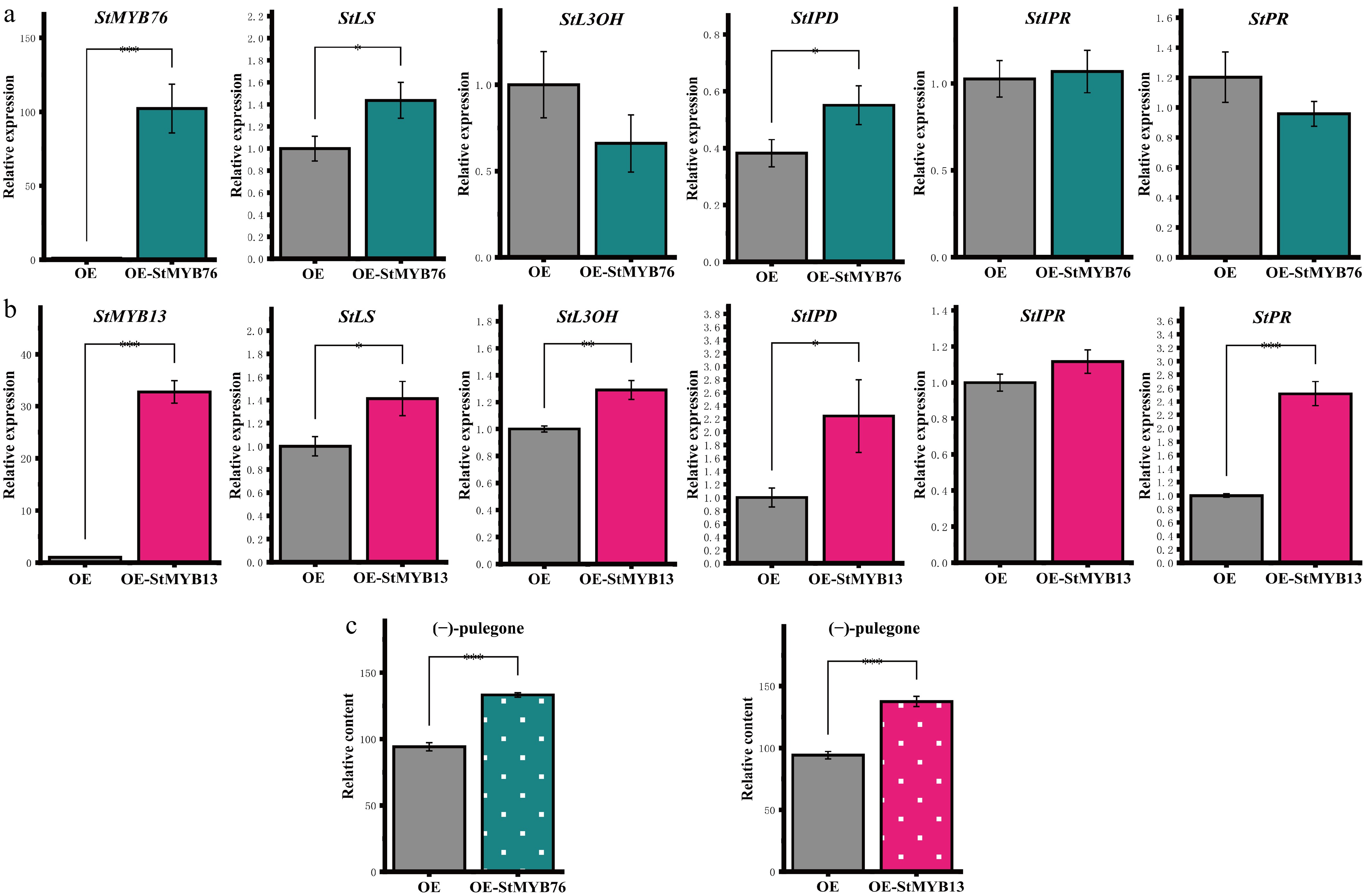
Figure 4.
Regulatory effects of StMYBs overexpression on the (−)-pulegone biosynthesis pathway. (a), (b) The figure illustrates the changes in the expression levels of StMYB76/StMYB13 and critical genes within the (−)-pulegone biosynthesis pathway. (c) The overexpression of StMYBs leads to changes in the concentration of (−)-pulegone monoterpene in S. tenuifolia. * p < 0.05, ** p < 0.01, *** p < 0.001, 'ns' denotes not significant.
-
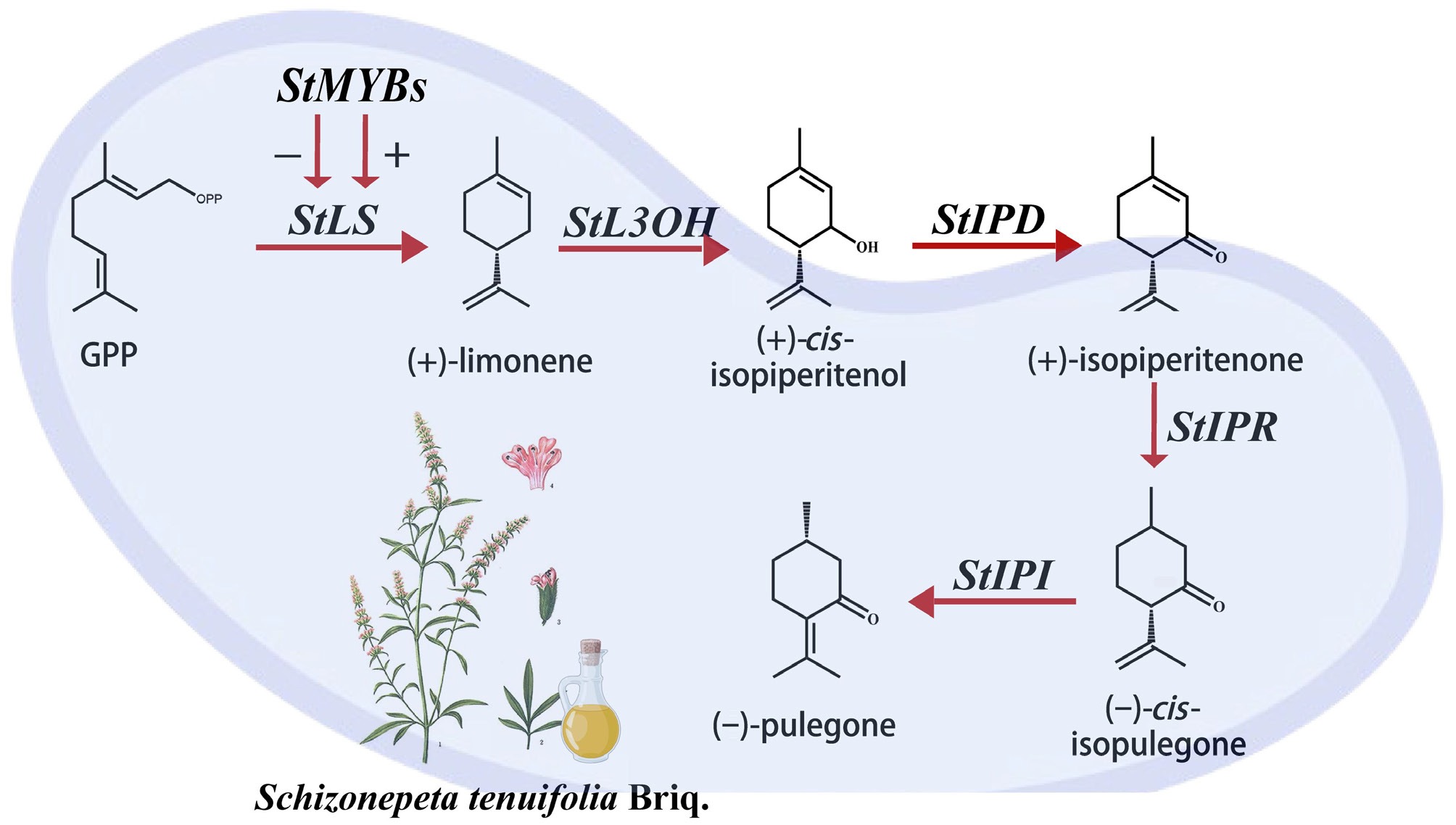
Figure 5.
StMYBs TFs influence S. tenuifolia. (−)-pulegone monoterpene biosynthesis mechanism map.
Figures
(5)
Tables
(0)