-
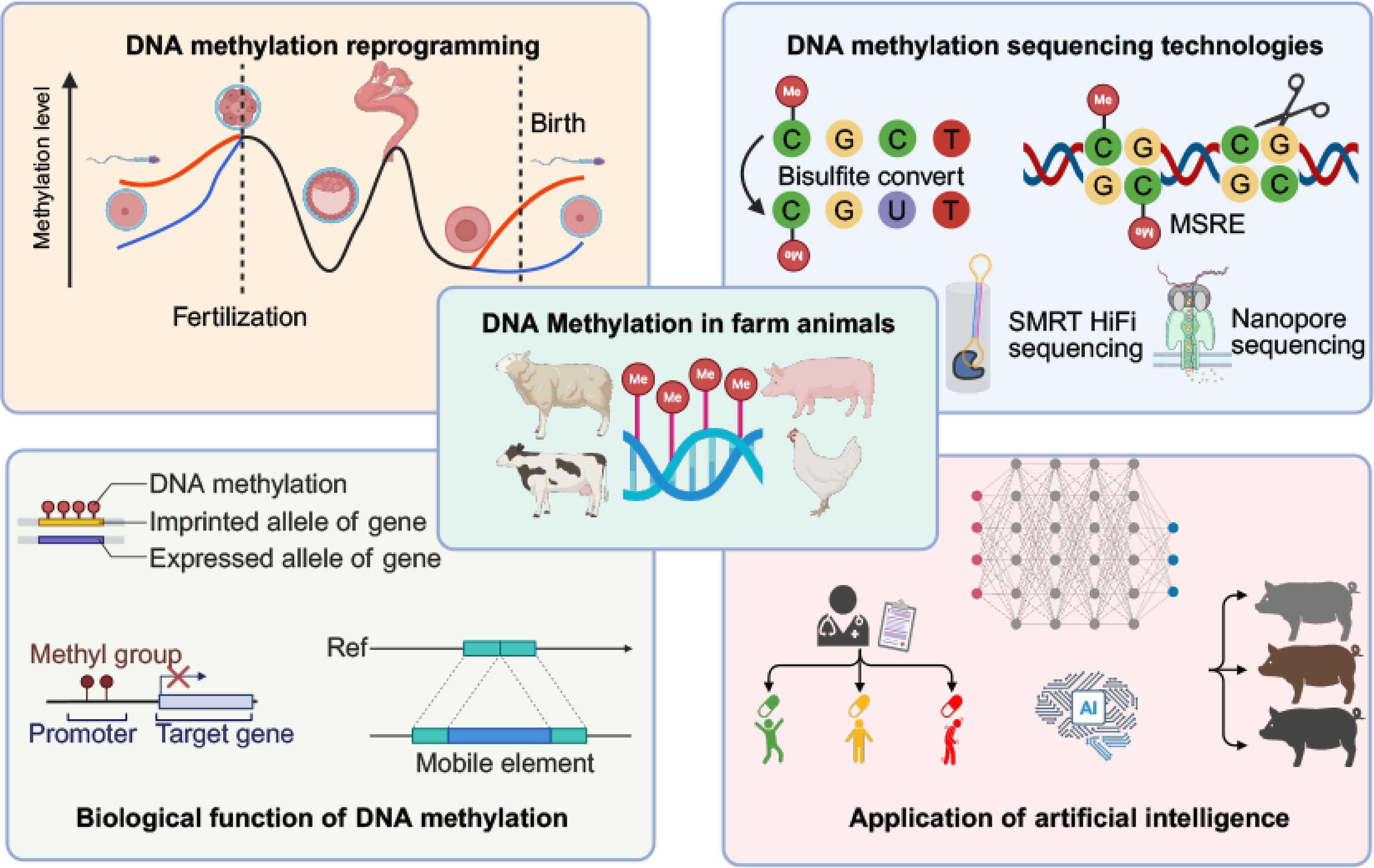
Figure 1.
Emerging frontiers of DNA methylation in livestock. MSRE: Methylation-sensitive restriction enzyme method.
-
Breeds Year Tissues Methods Findings Duroc[92] 2015 Fat, heart, kidney, liver, lung, lymph node, muscle, and spleen WGBS Adult pig methylomes closely resemble human, supporting biomedical relevance Landrace pigs[13] 2021 Longissimus dorsi WGBS Dynamic DNA methylation coordinates TF access and gene programs to drive porcine skeletal muscle development Ross308 chicken[99] 2021 Jejunum, ileum, breast muscle, spleen WGBS A multi-tissue WGBS-based methylation clock predicts broiler age and health Holstein cows[100] 2020 Mammary glands, whole blood cells, prefrontal cortex of the brain, and semen straws WGBS Multi-tissue methylation analyses show global and tissue-specific methylation patterns Meishan and Duroc[101] 2022 Testes MeDIP-seq LDHC promoter demethylation activates expression during porcine testis maturation F1 crossbreeds of Korean native and Yorkshire breeds[102] 2022 Abdominal fats MBD-seq Nanopore methylomes identify cattle age-related DMRs and pathways Holstein cows[103] 2023 Whole blood cells ONT Low-pass nanopore enables accurate genomic prediction plus simultaneous methylation profiling Huxu chicken[104] 2023 Muscle ONT Dot chromosomes and the W chromosome are hypermethylated, whereas centromere cores are relatively hypomethylated in Huxu chicken Göttingen minipigs[105] 2022 Whole blood cells ONT Nanopore cfDNA profiling in minipigs found 1,236 obesity DMRs, implicating PPARGC1B and metabolic pathways Brahman, Droughtmaster, and Tropical Composite[106] 2021 Tail hair ONT Portable ONT tail-hair CpG methylation profiles enabled a cattle epigenetic clock predicting age with ~0.71 correlation Note: Representative studies of different sequencing methods. Table 1.
Application of high-throughput methylation detection technology in livestock.
Figures
(1)
Tables
(1)