-
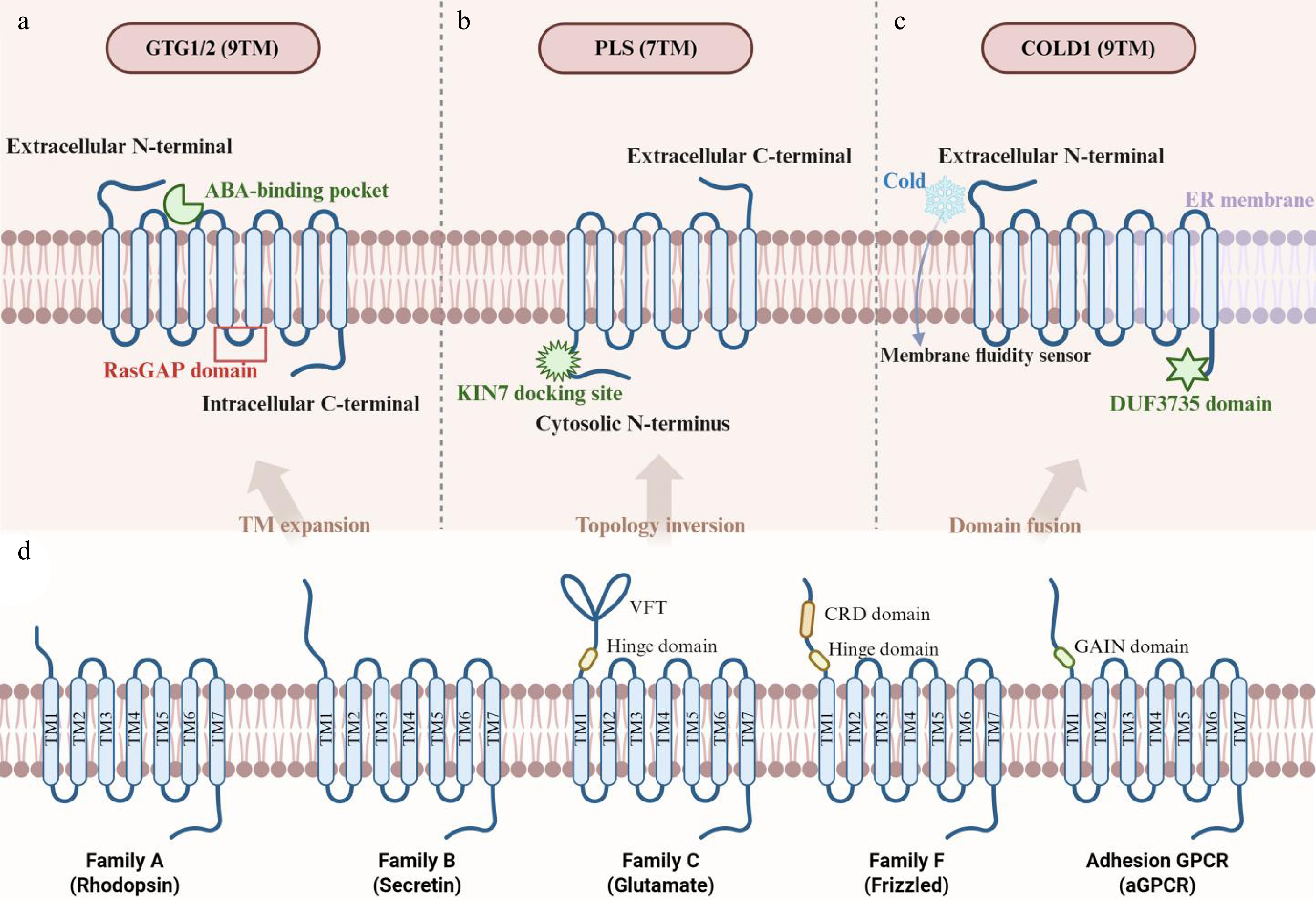
Figure 1.
Structural diversity of plant GPCR-like receptors compared to animal GPCRs. Protein structures are depicted schematically, highlighting transmembrane topology and key functional domains. The plasma membrane is represented by a light pink band. (a) GTG1/2 (9TM topology): illustrates a 9TM topology with an extracellular N-terminus (N). The third intracellular loop (ICL3) contains a degenerate Ras GTPase-activating protein (GAP) domain, critical for its intrinsic GTPase activity and stereoselective abscisic acid (ABA) binding within the transmembrane core. (b) PLS (7TM topology): depicts a 7TM topology with a distinctive cytosolic N-terminus (N), which interacts with adapter proteins like KIN7. This topology facilitates integration into the PRR-KIN7-PLS complex for pathogen signal perception. (c) COLD1 (9TM topology): shows a 9TM topology localized to the plasma membrane and endoplasmic reticulum (ER), featuring a distinctive domain of unknown function 3735 (DUF3735) domain of unknown function. This structure is proposed to sense cold-induced changes in membrane fluidity. (d) Canonical animal GPCR (7TM topology): serves as a reference, exemplifying the classical 7TM topology with an extracellular N-terminus, intracellular C-terminus, and conserved motifs for G protein coupling, following the traditional five-class classification system[22].
-
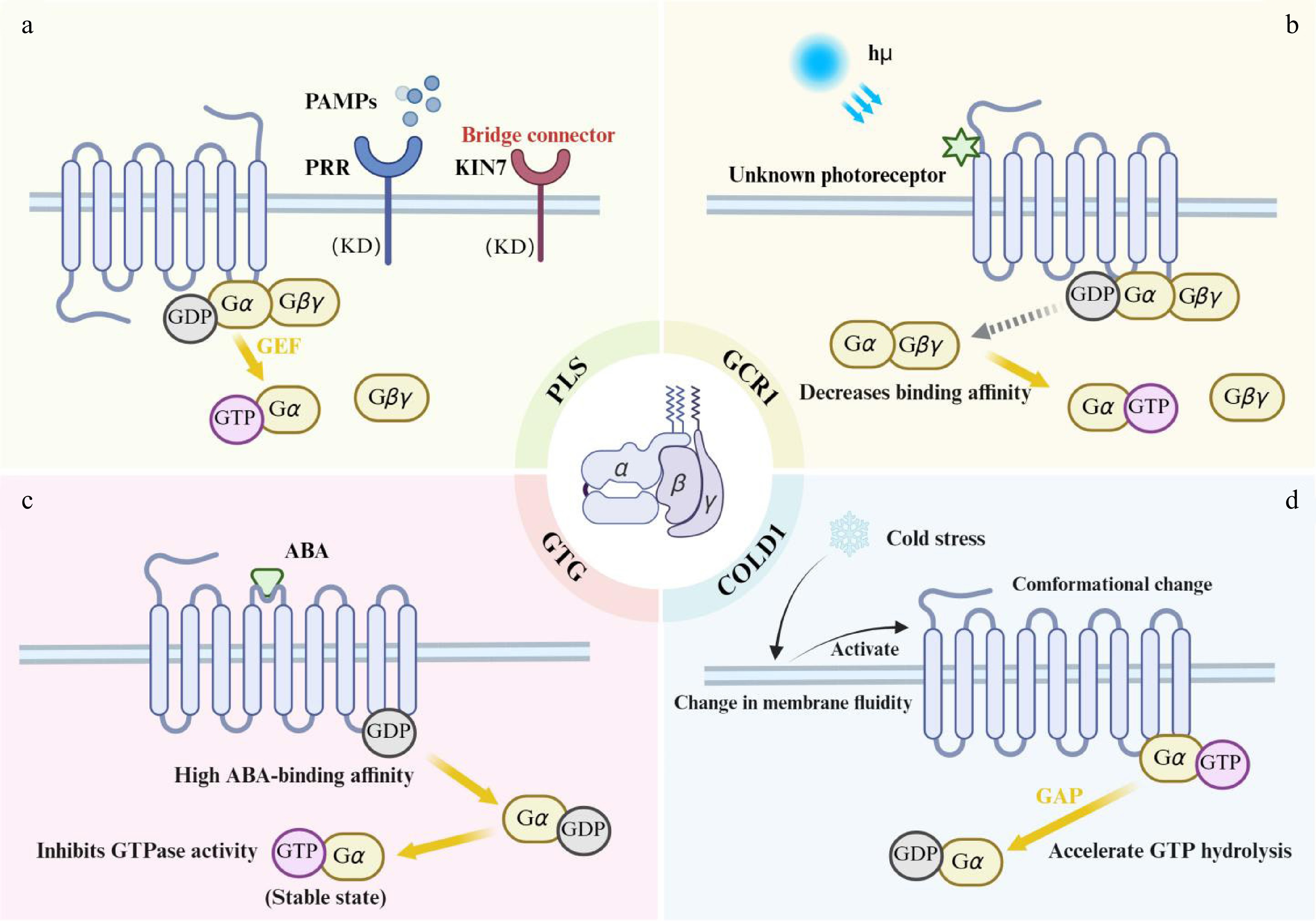
Figure 2.
G protein regulatory mechanisms of plant atypical GPCRs. (a) PLS (PAQR-like sensor): pathogen-associated molecular patterns (PAMPs) are perceived by pattern recognition receptors (PRRs). This leads to the formation of a PRR-KIN7-PLS ternary complex, which activates the guanine nucleotide exchange factor (GEF) activity of PLS. Activated PLS catalyzes GDP/GTP exchange on the Gα subunit. (b) GCR1: blue light activates GCR1 via an unidentified photoreceptor. The intracellular domain of activated GCR1 binds to GDP-bound Gα, reducing Gα's affinity for GDP and promoting GDP release. (c) GTG1/2: ABA binds stereoselectively to the high-affinity GDP-bound state of GTG. This triggers GPA1 binding, which inhibits the GTPase activity of GTG, thereby stabilizing the GTP-bound GTG state. (d) COLD1: cold stress increases membrane lipid rigidity, inducing a conformational change in COLD1. Acting as a GAP, COLD1 binds to GTP-bound Gα and accelerates GTP hydrolysis, rapidly generating GDP-bound Gα.
-
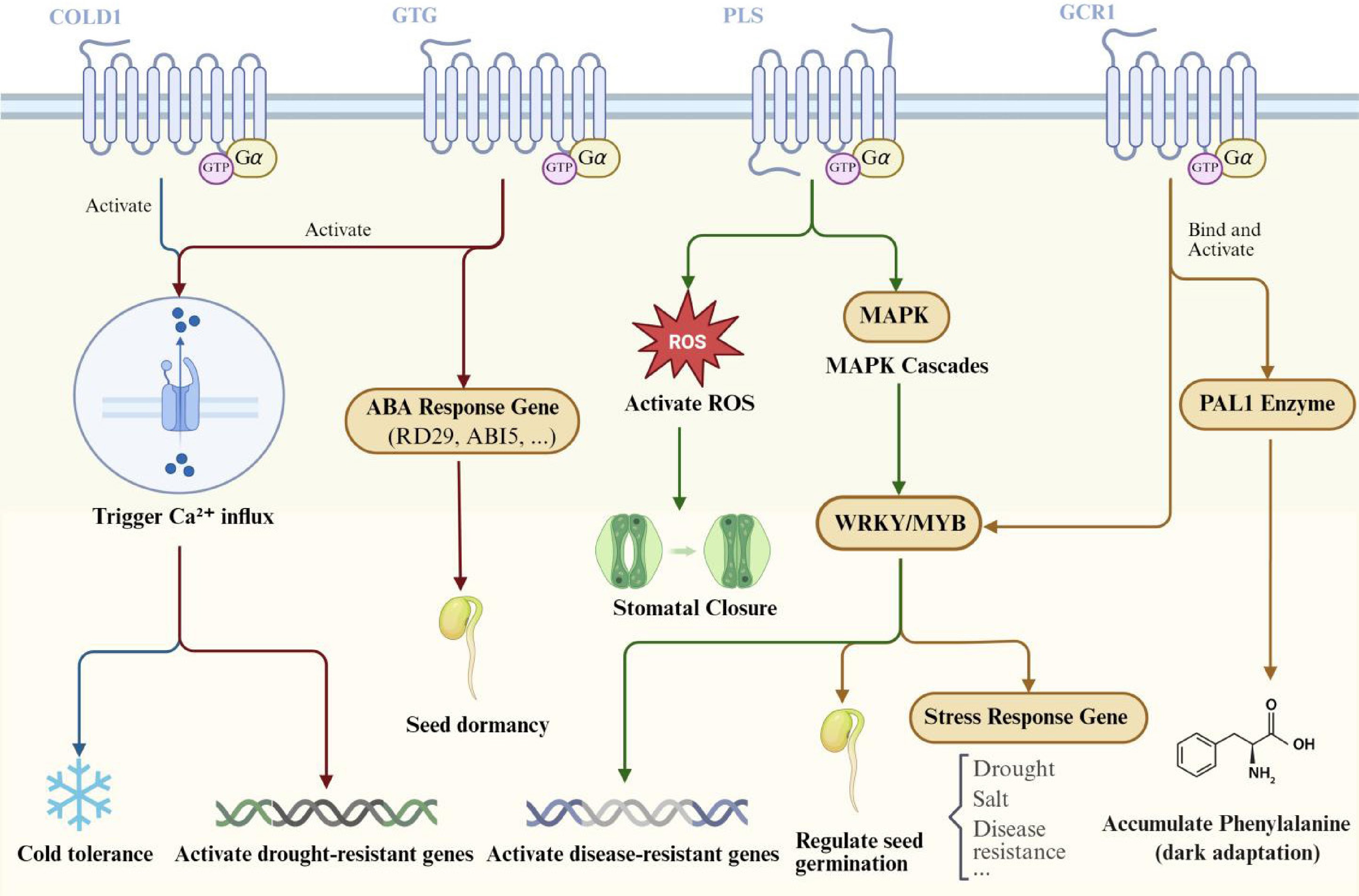
Figure 3.
Plant GPCR signaling pathways and their roles in environmental adaptation and developmental regulation. This schematic model delineates the downstream signaling pathways and associated physiological functions mediated by plant GPCRs, as detailed in the main text. COLD1-mediated cold sensing: cold stress induces membrane rigidification, leading to COLD1 activation. COLD1 functions as a GAP for RGA1 (Gα), accelerating GTP hydrolysis. This activation subsequently triggers Ca2+ influx, ultimately enhancing chilling tolerance. GTG1/2-mediated ABA Signaling: ABA binding to the GDP-bound state of GTG1/2 recruits GPA1 (Gα) and inhibits its GTPase activity, thereby stabilizing the active GTG-GTP state. This pathway regulates two primary outputs: (1) activation of plasma membrane Ca2+ channels and induction of drought-tolerance gene expression, and (2) activation of ABA-responsive genes to promote seed dormancy. PLS-mediated Immunity: Upon perception of pathogens by pattern recognition receptors (PRRs) or damage-associated molecular pattern (DAMP) receptors, the adaptor protein KIN7 is activated, initiating the PLS signaling pathway. PLS exhibits guanine nucleotide exchange factor (GEF) activity towards GPA1 (Gα), promoting GDP/GTP exchange. The activated G protein triggers a defense cascade comprising: a reactive oxygen species (ROS) burst, promotion of stomatal closure, and activation of a MAPK cascade, which in turn activates WRKY transcription factors to induce disease-resistance gene expression. GCR1-mediated Signaling: Activated by an unidentified blue light photoreceptor, GCR1 facilitates GTP loading of GPA1 (Gα). This process directly stimulates phenylalanine ammonia-lyase 1 (PAL1) enzyme activity, promoting phenylalanine synthesis. In parallel, the activated G protein regulates transcription factors such as WRKY to modulate seed germination and stress-responsive gene expression.
-
Feature Animal GPCRs Plant GPCR-like receptors Ref. Canonical topology 7TM 7TM or 9TM [13,21] Sequence motifs Conserved DRY, NPxxY / [2,18] Ligand perception Direct, high-affinity binding to single ligands Diverse: direct binding, indirect relay via complexes, mechano-sensing [14,19,21] GEF activity Ligand binding directly activates definitive GEF function Controversial/rare; often complex-dependent (PLS) or absent (GTG, COLD1) [13,14,18,21] G protein regulation Classical cycle: GPCR (GEF) → Gα-GTP → Effectors → GTP hydrolysis Diverse mechanisms: GEF (PLS), GAP (COLD1), GAP antagonism (GTG) [10,21] Table 1.
Comparative analysis of animal GPCRs and plant GPCR-like receptors.
-
GPCR candidate Species Topology Localization Ligand/signal perception G protein interaction Biological functions Ref. GCR1 A. thaliana 7TM Plasma membrane • Blue light (via unknown photoreceptor)
• ABA (controversial)• Interacts with GPA1
• Putative GEF (controversial)• Blue-light-induced Phe synthesis
• Seed germination
• Stress responses (drought, salt)[13,15,16,25] GTG1/GTG2 A. thaliana 9TM Plasma membrane • ABA (stereoselective) • GPA1 binding inhibits GTPase
• Acts as GAP antagonist• Stomatal closure
• Seed dormancy
• Drought response[28] COLD1 (OsCOLD1) O. sativa 9TM PM + ER • Cold stress (via membrane fluidity changes) • GAP for RGA1 • Chilling tolerance [29] PLS A. thaliana, O. sativa, G. max 7TM Plasma membrane • Pathogen signals (via PRR-KIN7)
• Damage signals (eATP, via P2K1)• Induced GEF activity
• Requires KIN7 adaptor• PTI immunity
• Stomatal defense
•Bacterial/fungal resistance
• MAPK activation
• ROS burst[23] PsGPCR P. sativum 7TM Plasma membrane (inferred) • Salt stress
• Heat stress• Interacts with pea Gα • Salinity/heat tolerance [24] LjGCR1 L. japonicus 7TM (inferred) Plasma membrane • Symbiotic nodulation signals • Interacts with G proteins (inferred) • Root nodule formation [25] ShGPCR1 S. officinarum 9TM (predicted) Plasma membrane • Membrane tension changes: Drought, Salt, Cold • Interacts with G proteins (inferred) • Multi-stress tolerance (drought, salt, cold)
• Activates stress-related genes
• Enhances antioxidant enzyme activity[34] Cand2/Cand7 A. thaliana 7TM (inferred) / • Bacterial AHLs (inferred) • Interacts with GPA1 • Rhizosphere communication
• Root development[45] TOM1 Gossypium spp. 7TM Plasma membrane (inferred) • Drought
• Cold• Interacts with GPA1 (inferred) • Drought/cold stress response [27] MLO O. sativa, A. thaliana, H. Vulgare 7TM Plasma membrane • Powdery mildew pathogens • Likely G protein-independent • Powdery mildew susceptibility [14] ZmCOLD1 Z. mays 9TM (predicted) PM + ER • Cold stress
• ABA signaling (inferred)• Interacts with Gα • Plant height regulation
• Chilling tolerance
• ABA responses[30,31] VaCOLD1 V. amurensis 9TM (predicted) PM + ER • Cold stress • Interacts with VaGPA1 • Cold stress tolerance [33] TaCOLD1 T. aestivum 9TM (predicted) PM + ER • Light signals
• Cold stress (inferred)• Interacts with TaGα-7A • Plant height regulation [32] Topology and localization marked with 'predicted' or 'inferred' are based on bioinformatic predictions or indirect evidence, not direct experimental validation. G protein interactions and ligand perceptions labeled as 'inferred' or 'putative' require further biochemical confirmation. Guanine nucleotide exchange factor (GEF) activity for most plant GPCR candidates remains controversial or unconfirmed. Table 2.
Summary of plant G protein-coupled receptor (GPCR) candidates.
-
Feature Animal GPCRs Plant GPCR candidates Ref. Canonical topology • 7TM domains
• Extracellular N-terminus
• Intracellular C-terminus
• Conserved across speciesDiverse architectures:
• 7TM (GCR1, PLS, PsGPCR, LjGCR1, Cand2, TOM1)
• 9TM (GTG1/2, COLD1,ZmCOLD1, ShGPCR1)
• Atypical orientation: Cytosolic N-terminus (PLS)[12,18,19,21,23−25,31,34] Sequence homology High conservation
• DRY motif
• NPxxY motifLow/no homology
• Absence of DRY/NPxxY motifs
• Limited similarity to animal GPCRs[2,18] Ligand perception Single-ligand focused
• Hormones, neurotransmitters
• Direct binding via N-terminusMultiligand integration
• GTG1/2: ABA + environmental cues
• PLS: Pathogens + damage signals via PRR-KIN7 complex
• ShGPCR1: Cold/salt/osmotic stress via membrane tension[14,22,23,28,34] Binding mechanisms Direct ligand binding
• Stereospecific pockets
• Ligand-induced conformational changesDiverse mechanisms:
• Hydrophobic pockets (GTG1/2)
• Membrane fluidity sensing (COLD1, ShGPCR1)
• Indirect relay (PLS: requires PRR/KIN7)
• GTP-state modulation (GTG1/2: GDP-bound state enhances ABA affinity)[14,19,23,28,29,34] GEF activity Definitive GEF function
• Catalyzes GDP→GTP exchange on Gα
• Triggers Gαβγ dissociationControversial/atypical:
• PLS: Induced GEF activity in PRR-KIN7 complex
• GCR1: Putative GEF (no direct evidence)
• Absent in GTG/COLD families[12,18,19,23,28,29] G protein regulation Classical cycle:
1. GPCR-GEF activates Gα
2. Gα-GTP dissociates from Gβγ
3. GTP hydrolysis resets systemDiverse mechanisms:
• GAP activity (COLD1: accelerates GTP hydrolysis)
• GTPase antagonism (GTG1/2: GPA1 inhibits GTPase)
• Direct effector modulation (Gβγ regulates Ca2+ channels independently)
• Kinase-dependent (PLS: requires P2K1/KIN7 phosphorylation)[14,21,28,29] Key domains • Ligand-binding domains
• G-protein coupling domainsNovel domains:
• RasGAP domain (GTG1/2)
• DUF3735 (ZmCOLD1)
• GTP-binding domains (ShGPCR1, GTG1/2)[14,19,28,33,34] Subcellular localization Plasma membrane Dual localization:
• Plasma membrane + ER (COLD1, ZmCOLD1)
• Plasma membrane mostly (others)[13,21,28,29,33] Signaling cascades • cAMP/PKA
• Ca2+ mobilization
• MAPK activationPlant-specific pathways:
• Ca2+ signaling hubs (COLD1, ShGPCR1)
• MAPK immunity cascade (PLS)
• Transcriptional networks (GCR1, GTG1/2)[13,19,21,23,28,34,42] Evolutionary innovations Conserved 7TM architecture Structural innovations:
• 9TM topology (GTG/COLD families)
• Cytosolic N-terminus (PLS)
• Functional domain fusion (COLD1: ER localization + Ca2+ regulation)
• Mechanosensing (ShGPCR1: membrane tension transduction)[19,21,23,28,29,34] Controversies Well-established paradigm Ongoing debates:
• Existence of bona fide GPCRs in plants
• GCR1/GCR2 classification (GCR2 reclassified as LanC-like protein)
• GPA1 self-activation vs. GPCR-dependence[2,17,18] Table 3.
Comparative analysis of animal and plant G protein-coupled receptors.
Figures
(3)
Tables
(3)