-
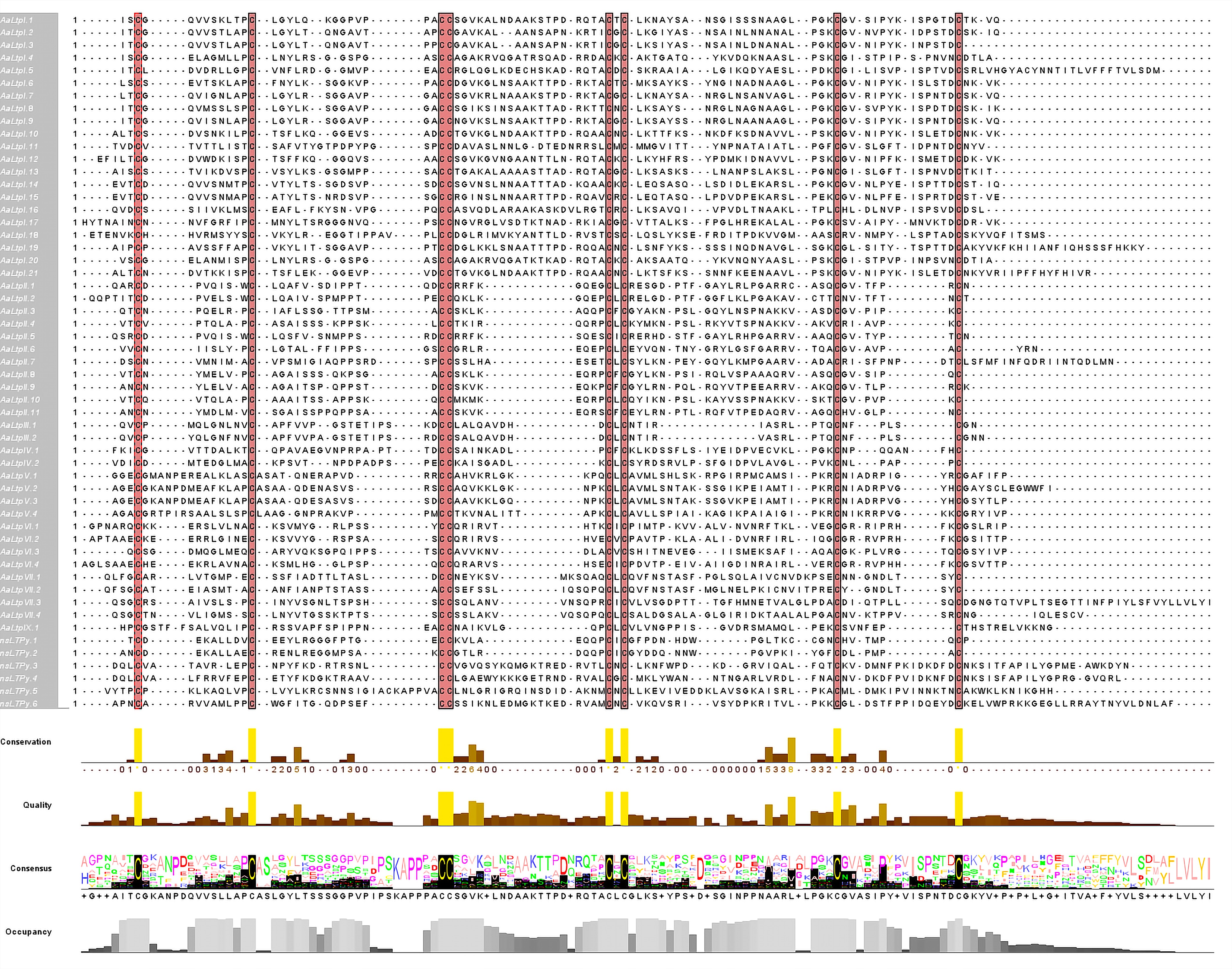
Figure 1.
Alignment of multiple sequences of AaLTP proteins. The conserved cysteine residues are marked against red backgrounds.
-
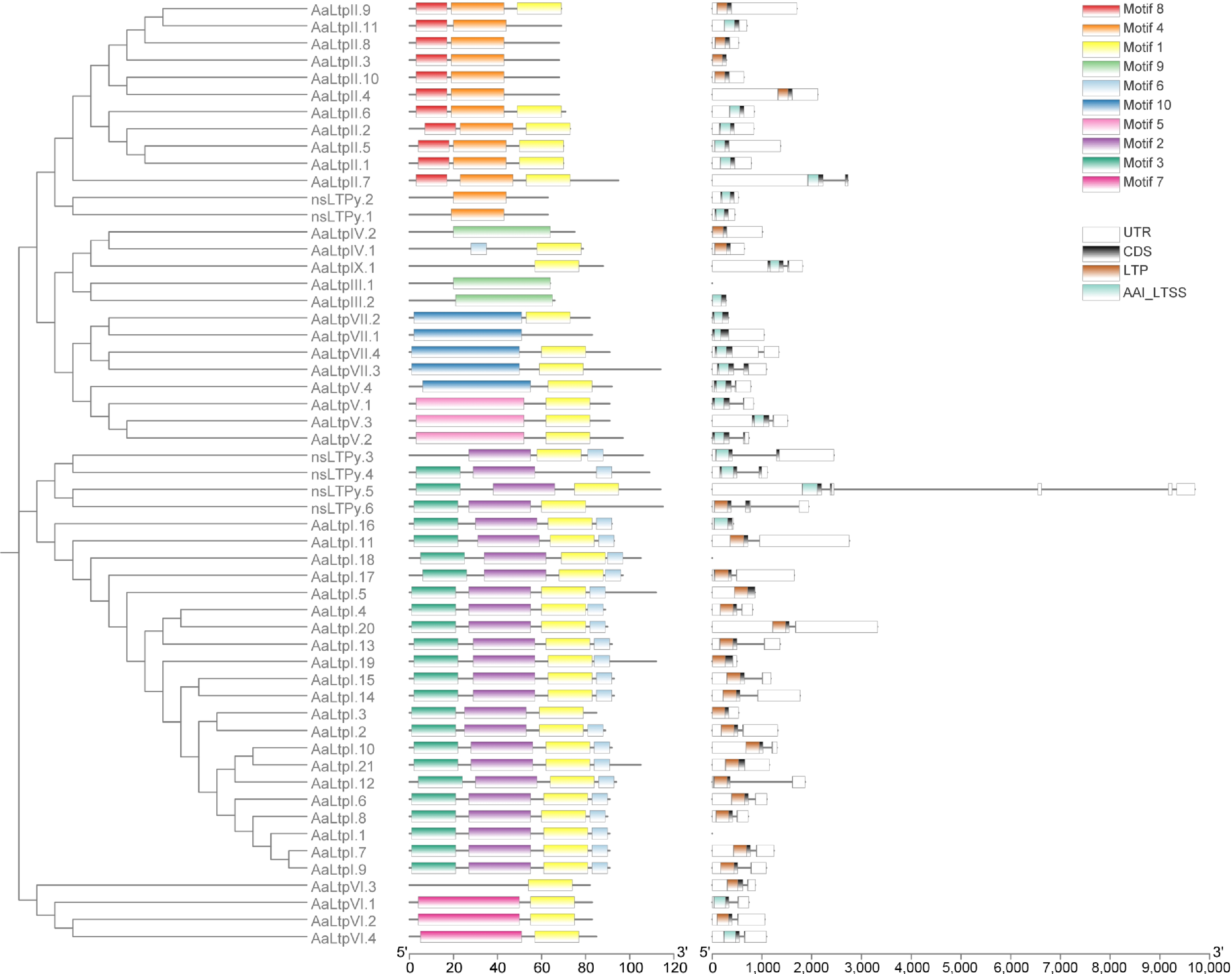
Figure 2.
Phylogenetic tree of AaLTPs together with motif analysis and gene structure analysis. The full length of mature protein sequences of nsLTPs from A. annua was used to construct the phylogenetic tree using a Maximum likelihood method. Motif analysis was performed using MEME, while domain analysis was performed using the batch Web CD-Search tool. All of the above results were visualized together with the gene annotation file.
-
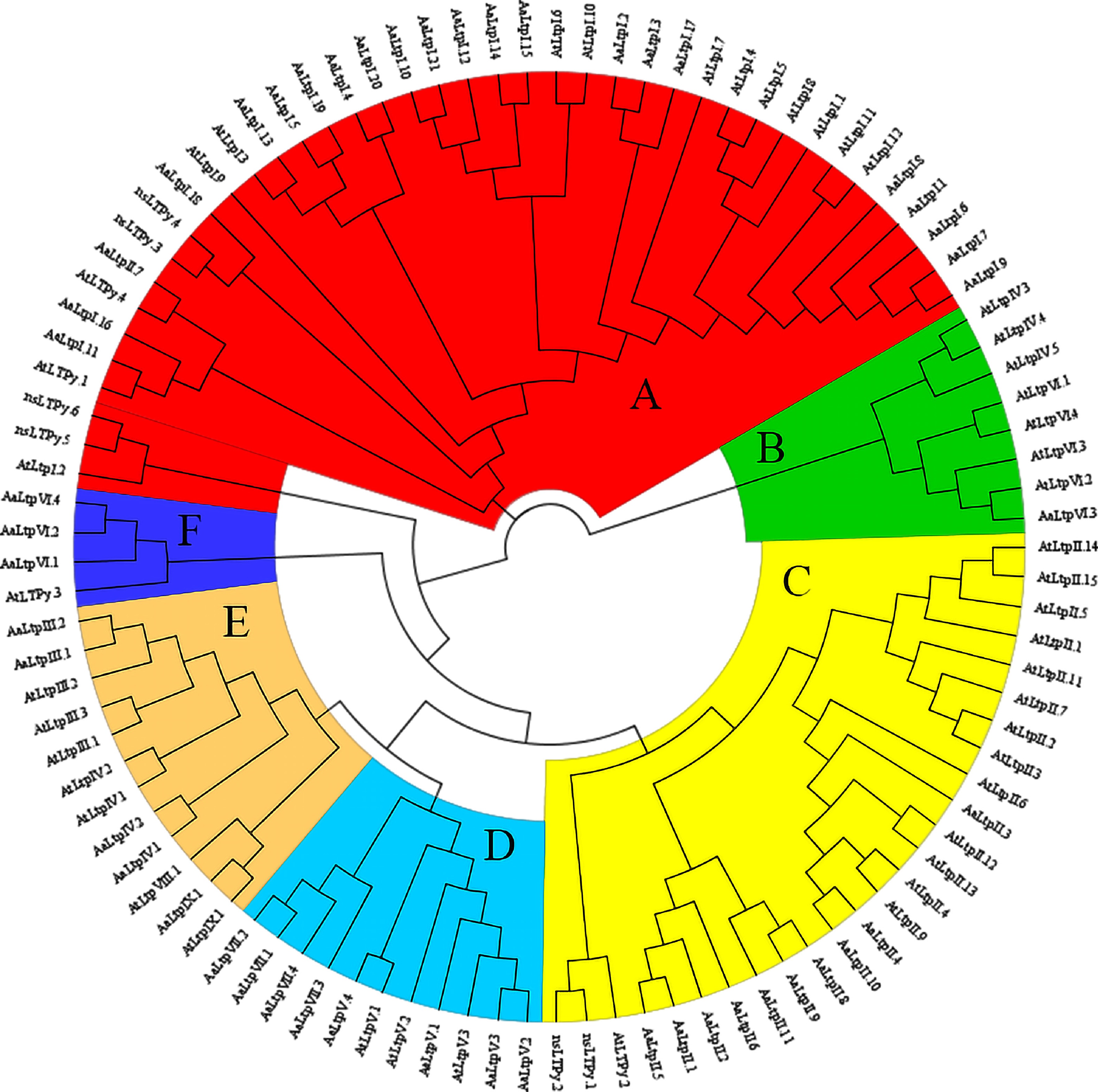
Figure 3.
Phylogenetic tree of nsLTPs in A. thaliana and A. annua. The full length of mature protein sequences of nsLTPs from A. thaliana and A. annua were used to construct the phylogenetic tree using a Maximum likelihood method. Different colors represent different clusters.
-
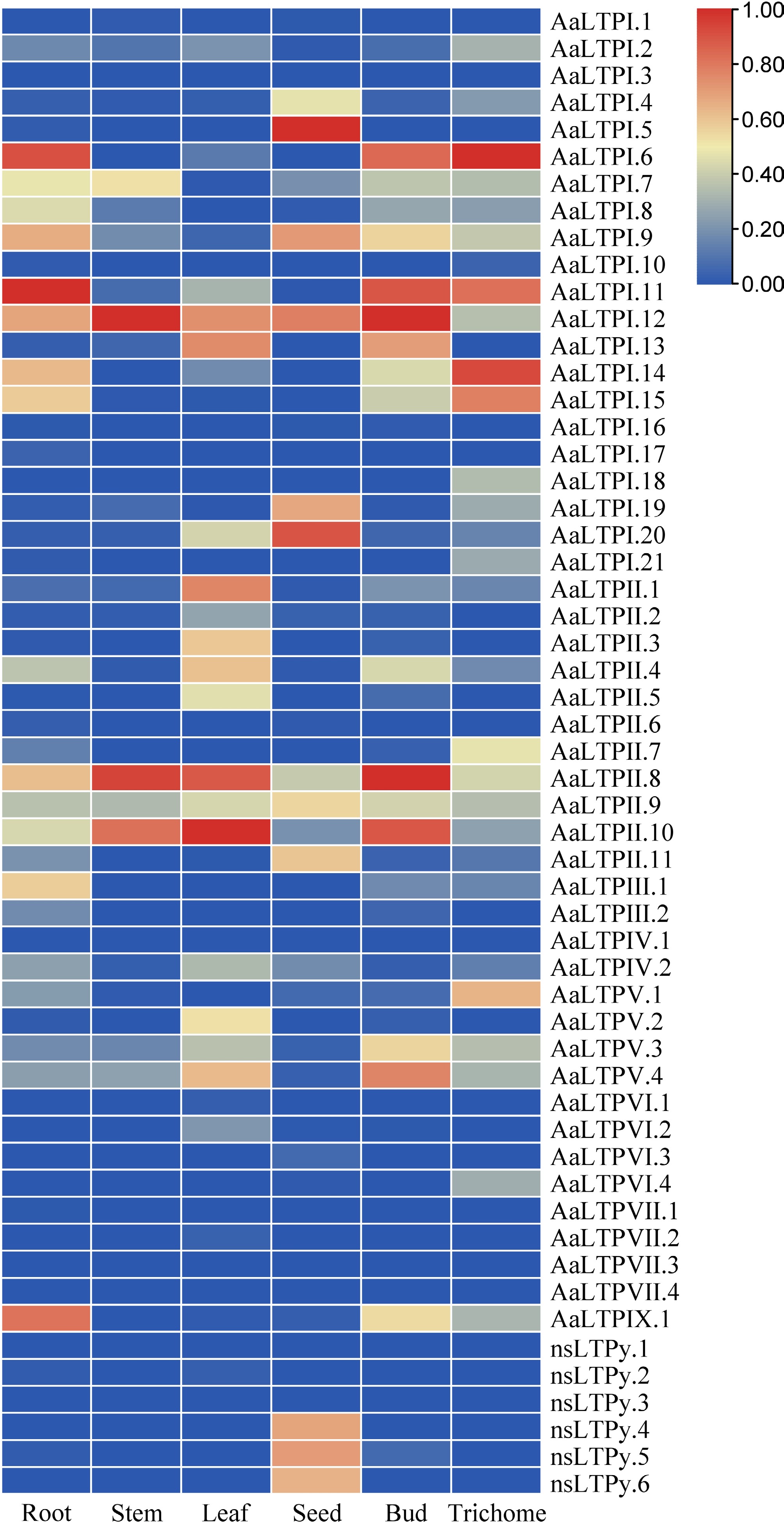
Figure 4.
Expression analysis of AaLTPs. The heatmap shows expression comparison between six organs of AaLTPs. The data shown were the results of column scaling each gene by RNA-seq data using the Zero To One Scale Method. The color bar represents the relative expression level.
-
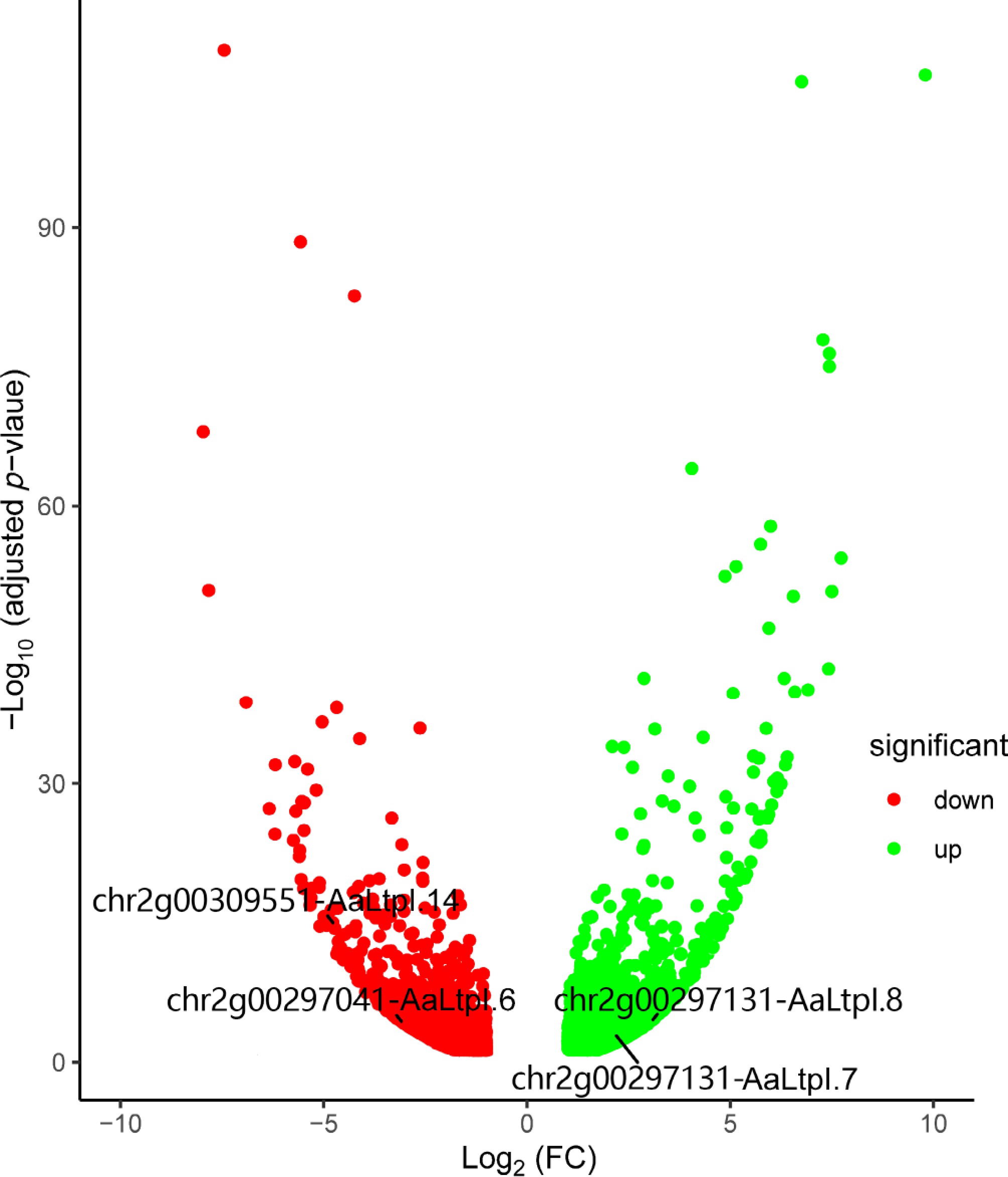
Figure 5.
Volcano plot shows differentially expressed genes in AaHD8-RNAi. The red points indicate down-regulated genes, while the green ones indicate up-regulated genes. FC stands for fold change.
-
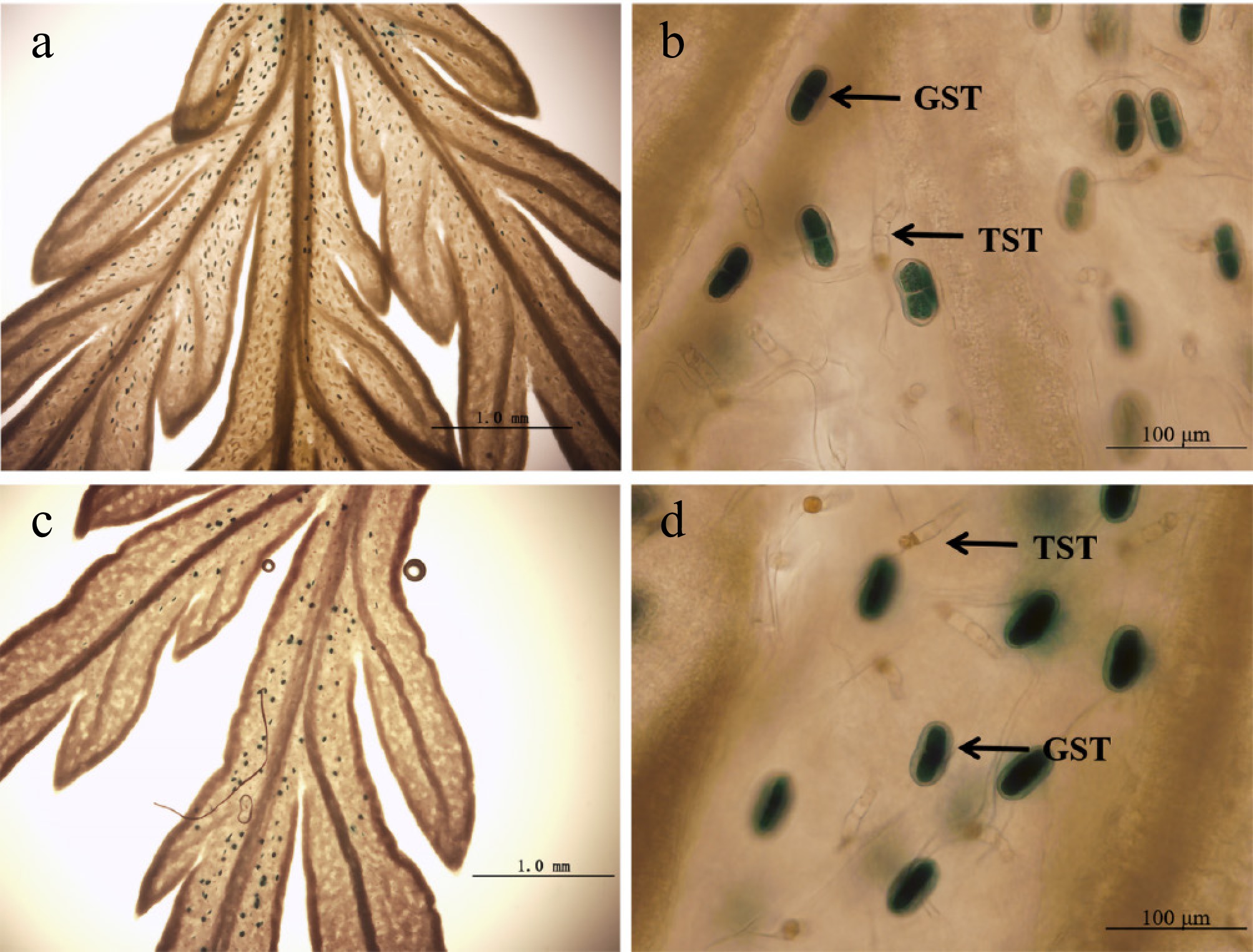
Figure 6.
GUS expression (blue) in A. annua plants transformed with the 1391Z-proLTP1-GUS and 1391Z-proLTP2-GUS vector. (a), (b) Leaves of 1391Z-proLTP1-GUS plants. (c), (d) Leaves of 1391Z-proLTP2-GUS plants.
-
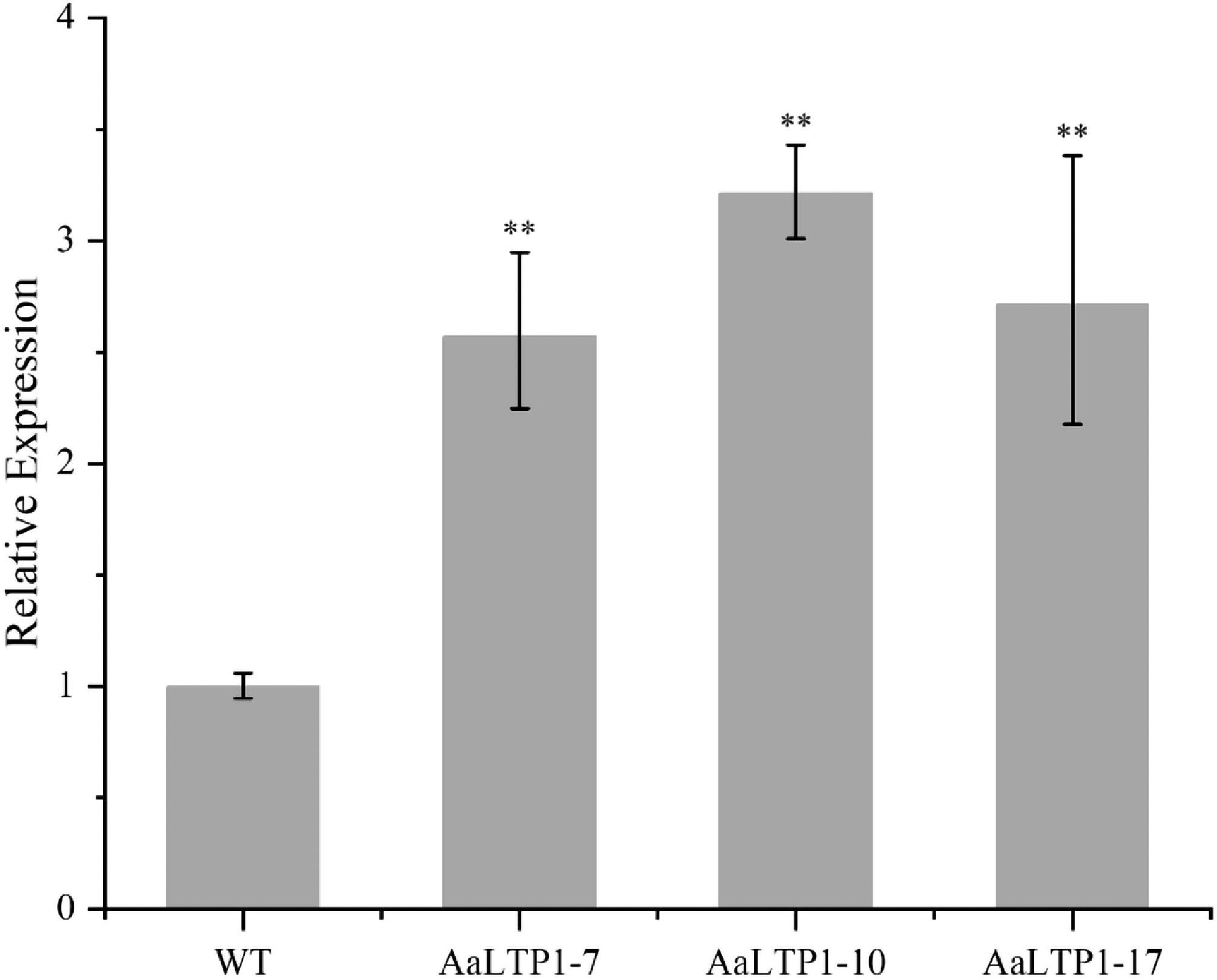
Figure 7.
Expression levels of AaLTP1 in AaLTP1 over-expression plants measured by qRT-PCR. The average expression level of genes in WT was set as 1. Actin was used as internal control; the error bars show means ± SD from three technical repeats; Student's t-test ** p < 0.01, * p < 0.05.
-
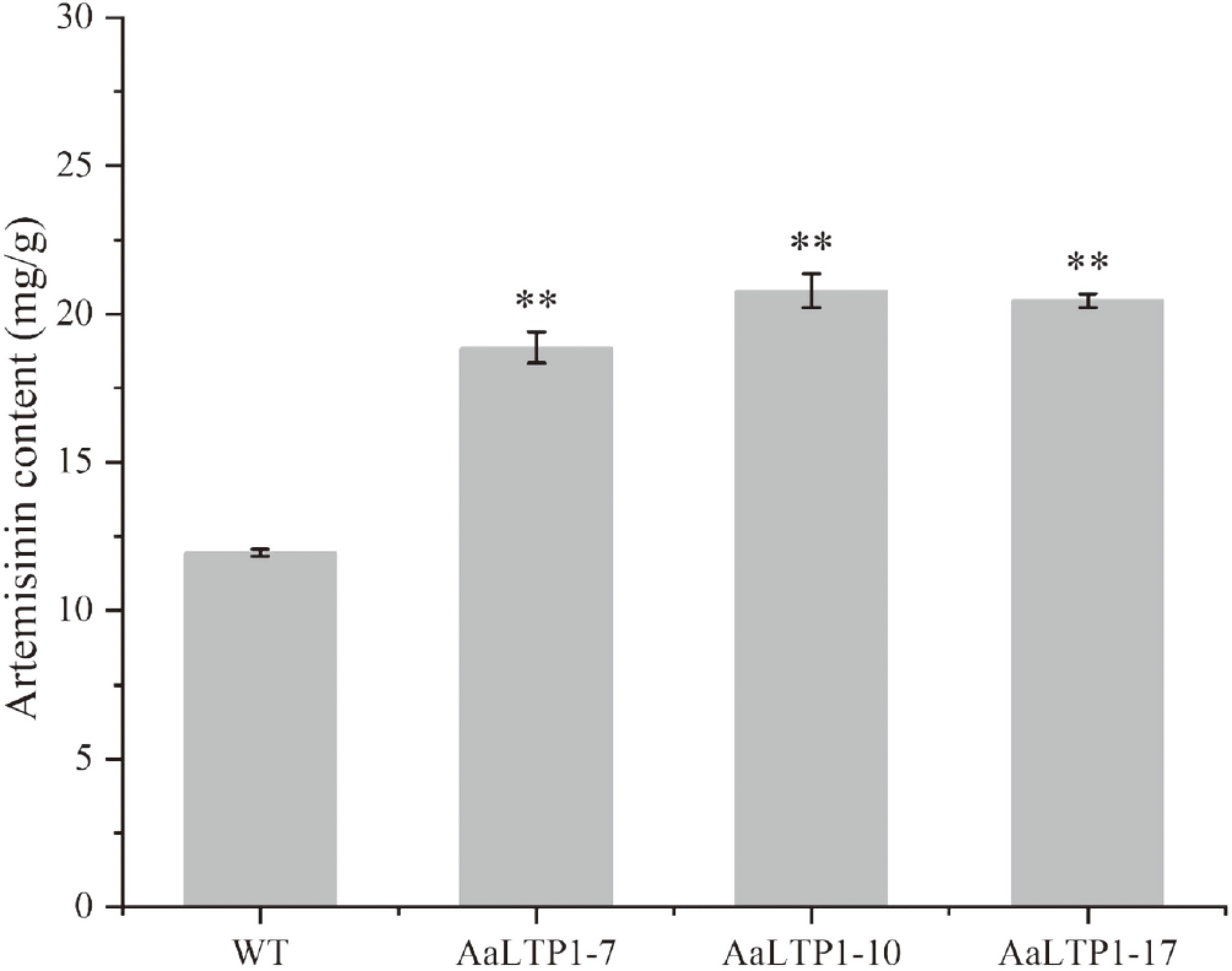
Figure 8.
Artemisinin contents in AaLTP1 over-expression lines were measured by HPLC. Error bars indicate ± SD of three biological replicates. ** p < 0.01, Student's t-test.
-
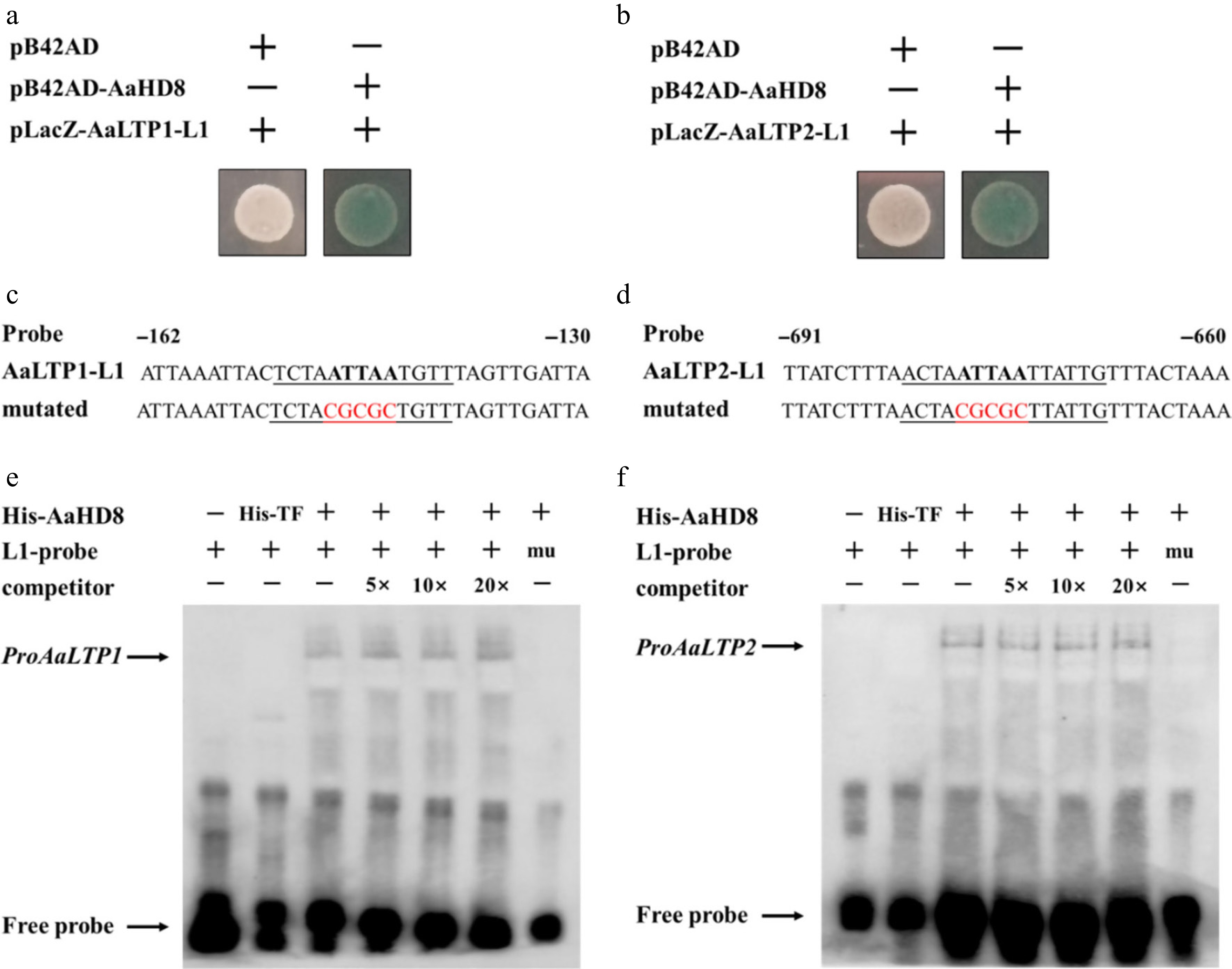
Figure 9.
AaHD8 directly binds to the promoter of AaLTP1 and AaLTP2. (a), (b) Yeast one-hybrid assay of protein–DNA interaction. pB42AD is an empty vector used as a negative control. (c), (d) Probe and mutant probe. (e), (f) AaHD8 binds to the L1-box-like motif from AaLTP; 5×, 10×, and 20× indicate the dilution multiple of cold competitors relative to that of the labeled probe; mu: labeled mutated L1-probes were tested as negative controls; His-TF protein was used as a negative control.
-
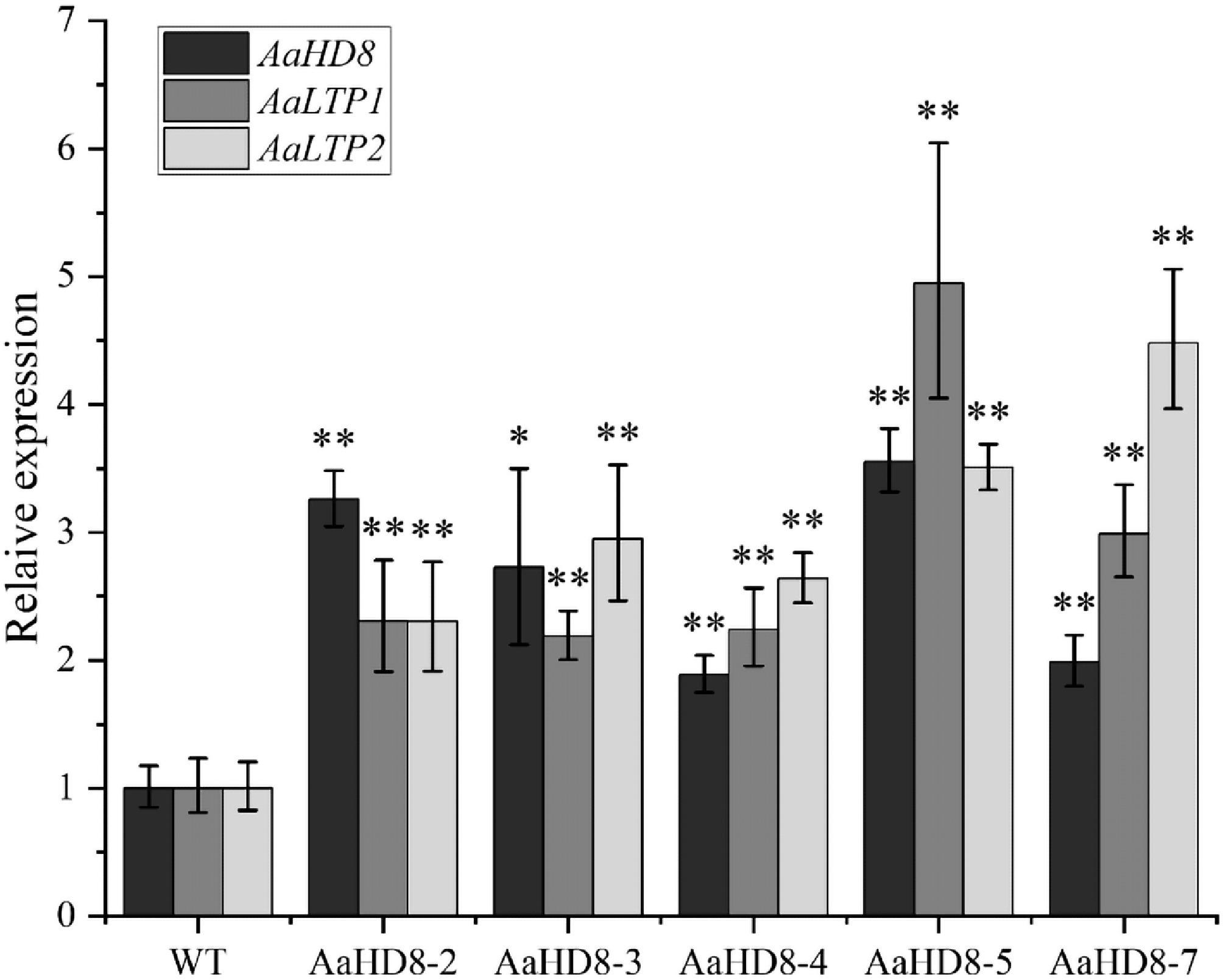
Figure 10.
Relative expression of AaHD8 and AaLTP in AaHD8 over-expression plants. The average expression level of genes in WT was set as 1. Actin was used as internal control; the error bars show means ± SD from three technical repeats; Student's t-test ** p < 0.01, * p < 0.05.
-
Name Gene ID CDS length (bp) AAa SPb SLc MPd (AA) ECMe MP (MWf) MP (pIg) Type I AaLtpI.1 Super-scaffold_
100123g01462191351 116 25 s 91 C-X9-C-X13-CC-X19-CXC-X22-C-X13-C 9,106.48 9 AaLtpI.2 chr2g00359671 345 114 25 s 89 C-X9-C-X13-CC-X17-CXC-X22-C-X13-C 9,016.44 8.87 AaLtpI.3 chr2g00359701 333 110 25 s 85 C-X9-C-X13-CC-X17-CXC-X22-C-X13-C 8,599.97 8.7 AaLtpI.4 chr3g00499441 345 114 25 s 89 C-X9-C-X13-CC-X19-CXC-X21-C-X12-C 9,076.39 9.05 AaLtpI.5 chr1g00152381 411 136 24 s 112 C-X9-C-X13-CC-X19-CXC-X21-C-X13-C 12,177.13 5.54 AaLtpI.6 chr2g00297041 351 116 25 s 91 C-X9-C-X13-CC-X19-CXC-X22-C-X13-C 9,439.93 9.19 AaLtpI.7 chr2g00297131 351 116 25 s 91 C-X9-C-X13-CC-X19-CXC-X22-C-X13-C 9,198.68 9.57 AaLtpI.8 chr2g00297171 348 115 25 s 90 C-X9-C-X13-CC-X19-CXC-X21-C-X13-C 9,019.42 9.26 AaLtpI.9 chr2g00297151 351 116 25 s 91 C-X9-C-X13-CC-X19-CXC-X22-C-X13-C 9,092.46 9.3 AaLtpI.10 chr2g00297111 354 117 25 s 92 C-X9-C-X13-CC-X19-CXC-X22-C-X13-C 9,763.21 8.66 AaLtpI.11 chr3g00546061 369 122 29 s 93 C-X9-C-X16-CC-X19-CXC-X21-C-X13-C 9,745.02 3.72 AaLtpI.12 chr2g00297121 342 113 19 s 94 C-X9-C-X13-CC-X19-CXC-X22-C-X13-C 10,230.87 8.67 AaLtpI.13 chr1g00017271 360 119 27 s 92 C-X9-C-X14-CC-X19-CXC-X21-C-X13-C 9,043.43 8.89 AaLtpI.14 chr2g00309551 354 117 24 s 93 C-X9-C-X14-CC-X19-CXC-X22-C-X13-C 9,682.74 4.14 AaLtpI.15 chr4g00612411 369 122 29 s 93 C-X9-C-X14-CC-X19-CXC-X22-C-X13-C 10,033.22 4.76 AaLtpI.16 chr6g01084791 363 120 28 s 92 C-X9-C-X14-CC-X20-CXC-X21-C-X13-C 9,795.49 7.69 AaLtpI.17 chr2g00297031 360 119 22 s 97 C-X9-C-X15-CC-X19-CXC-X22-C-X12-C 10,454.11 9.3 AaLtpI.18 unctg_3864g01516351 390 129 24 s 105 C-X9-C-X16-CC-X19-CXC-X23-C-X12-C 11,775.69 8.46 AaLtpI.19 chr2g00297011 414 137 25 s 112 C-X9-C-X14-CC-X19-CXC-X22-C-X12-C 11,964.69 9.09 AaLtpI.20 chr3g00499431 348 115 25 s 90 C-X9-C-X13-CC-X19-CXC-X21-C-X13-C 9,170.5 9.16 AaLtpI.21 chr2g00297081 393 130 25 s 105 C-X9-C-X13-CC-X19-CXC-X22-C-X13-C 11,565.33 8.65 Type II AaLtpII.1 chr1g00100031 294 97 27 s 70 C-X7-C-X13-CC-X8-CXC-X19-C-X3-C 7,771.9 8.47 AaLtpII.2 chr1g00099971 297 98 25 s 73 C-X7-C-X13-CC-X8-CXC-X20-C-X2-C 7,861.2 5.06 AaLtpII.3 chr1g00099961 285 94 26 s 68 C-X7-C-X13-CC-X8-CXC-X23-C-X6-C 7,196.39 8.88 AaLtpII.4 chr5g00892001 288 95 27 s 68 C-X7-C-X13-CC-X8-CXC-X23-C-X6-C 7,424.05 9.96 AaLtpII.5 chr1g00099991 294 97 27 s 70 C-X7-C-X13-CC-X8-CXC-X23-C-X6-C 7,999.08 8.95 AaLtpII.6 chr3g00577871 291 96 25 s 71 C-X7-C-X13-CC-X8-CXC-X23-C-X6-C 7,771.01 8.5 AaLtpII.7 chr6g01008841 363 120 25 s 95 C-X7-C-X16-CC-X9-CXC-X23-C-X9-C 10,504.09 4.75 AaLtpII.8 chr3g00577891 294 97 29 s 68 C-X7-C-X13-CC-X8-CXC-X23-C-X6-C 7,165.38 8.91 AaLtpII.9 chr9g01375721 294 97 28 s 69 C-X7-C-X13-CC-X8-CXC-X23-C-X6-C 7,633.87 8.89 AaLtpII.10 chr5g00891931 294 97 29 s 68 C-X7-C-X13-CC-X8-CXC-X23-C-X6-C 7,245.69 9.41 AaLtpII.11 chr3g00483861 300 99 30 s 69 C-X7-C-X14-CC-X8-CXC-X23-C-X6-C 7,522.63 7.74 Type III AaLtpIII.1 unctg_5142g01560471 276 91 27 s 64 C-X9-C-X16-CC-X9-CXC-X12-C-X5-C 6,821.95 5.43 AaLtpIII.2 chr3g00559411 282 93 27 s 66 C-X9-C-X17-CC-X9-CXC-X12-C-X5-C 7,061.03 5.43 Type IV AaLtpIV.1 chr2g00348561 321 106 27 s 79 C-X9-C-X17-CC-X9-CXC-X17-C-X6-C 8,505.87 6.7 AaLtpIV.2 chr8g01299581 294 97 22 s 75 C-X9-C-X15-CC-X9-CXC-X24-C-X6-C 7,822.12 4.34 Type V AaLtpV.1 chr1g00206751 363 120 29 s 91 C-X14-C-X14-CC-X11-CXC-X16-C-X7-C 9,915.79 9.57 AaLtpV.2 chr1g00206761 378 125 28 s 97 C-X14-C-X14-CC-X11-CXC-X24-C-X10-C 10,313.99 8.62 AaLtpV.3 chr3g00569011 357 118 27 s 91 C-X14-C-X14-CC-X11-CXC-X24-C-X10-C 9,379.82 8.18 AaLtpV.4 chr5g00787181 363 120 28 s 92 C-X14-C-X14-CC-X12-CXC-X24-C-X10-C 9,418.59 10.4 Type VI AaLtpVI.1 chr6g01040821 318 105 22 s 83 C-X10-C-X12-CC-X9-CXC-X22-C-X9-C 9,389.32 10.33 AaLtpVI.2 chr6g01040781 318 105 22 s 83 C-X10-C-X12-CC-X9-CXC-X22-C-X9-C 9,111.7 9.45 AaLtpVI.3 chr1g00008331 336 111 29 s 82 C-X10-C-X16-CC-X9-CXC-X22-C-X9-C 8,667.06 6.71 AaLtpVI.4 chr6g01040811 333 110 25 s 85 C-X10-C-X12-CC-X9-CXC-X23-C-X9-C 9,188.67 8.53 Type VII AaLtpVII.1 chr1g00063881 333 110 27 s 83 C-X9-C-X14-CC-X12-CXC-X18-C-X7-C 8,888.1 4.39 AaLtpVII.2 chr1g00063861 330 109 27 s 82 C-X9-C-X14-CC-X12-CXC-X18-C-X7-C 8,722.86 4.25 AaLtpVII.3 chr2g00408761 423 140 26 s 114 C-X9-C-X14-CC-X12-CXC-X25-C-X9-C 12,044.73 4.51 AaLtpVII.4 chr2g00415681 345 114 23 s 91 C-X9-C-X14-CC-X12-CXC-X26-C-X9-C 9,136.58 8.45 Type IX AaLtpIX.1 chr2g00241771 339 112 24 s 88 C-X13-C-X15-CC-X9-CXC-X22-C-X6-C 9,369.99 8.21 nsLTPy nsLTPy.1 chr4g00736531 270 89 26 s 63 C-X8-C-X13-CC-X8-CXC-X17-C-X6-C 6,862.86 4.59 nsLTPy.2 chr4g00736521 273 90 27 s 63 C-X8-C-X13-CC-X8-CXC-X17-C-X6-C 6,910.89 4.56 nsLTPy.3 chr2g00265921 399 132 26 s 106 C-X9-C-X12-CC-X19-CXC-X19-C-X14-C 12,314.22 8.44 nsLTPy.4 chr2g00265911 405 134 25 s 109 C-X10-C-X13-CC-X19-CXC-X21-C-X14-C 12,409.34 8.98 nsLTPy.5 chr4g00694091 423 140 26 s 114 C-X9-C-X22-CC-X19-CXC-X25-C-X14-C 12,533.12 9.44 nsLTPy.6 chr2g00265901 423 140 25 s 115 C-X9-C-X12-CC-X19-CXC-X21-C-X14-C 13,023.16 8.44 a AA, number of amino acids; b SP, signal peptide; c SL, subcellular location, s = secretory pathway; d MP, mature protein; e ECM, eight cysteine motif; f MW, molecular weight in Dalton; g pI, isoelectric point. The detailed classification basis is listed in Supplementary Table S4. Table 1.
Putative nsLTP genes identified in the genome of A. annua L.
-
nsLTP type ECM and number of flanking amino acid residues 1 2 3, 4 5 6 7 8 Type I X C X9 C X13–16 CC X17, 19, 20 C X C X21−23 C X12, 13 C X Type II X C X7 C X13, 14, 16 CC X8, 9 C X C X19, 20, 23 C X2, 3, 6, 9 C X Type III X C X9 C X16, 17 CC X9 C X C X12 C X5 C X Type IV X C X9 C X15, 17 CC X9 C X C X17, 24 C X6 C X Type V X C X14 C X14 CC X11, 12 C X C X16, 24 C X7, 10 C X Type VI X C X10 C X12, 16 CC X9 C X C X22, 23 C X9 C X Type VII X C X9 C X14 CC X12 C X C X18, 25, 26 C X7, 9 C X Type IX X C X13 C X15 CC X9 C X C X22 C X6 C X Type XI X C X9 C X18−20 CC X13 C X C X24, 25 C X9 C X Table 2.
Diversity of the eight cysteine motif.
Figures
(10)
Tables
(2)