-
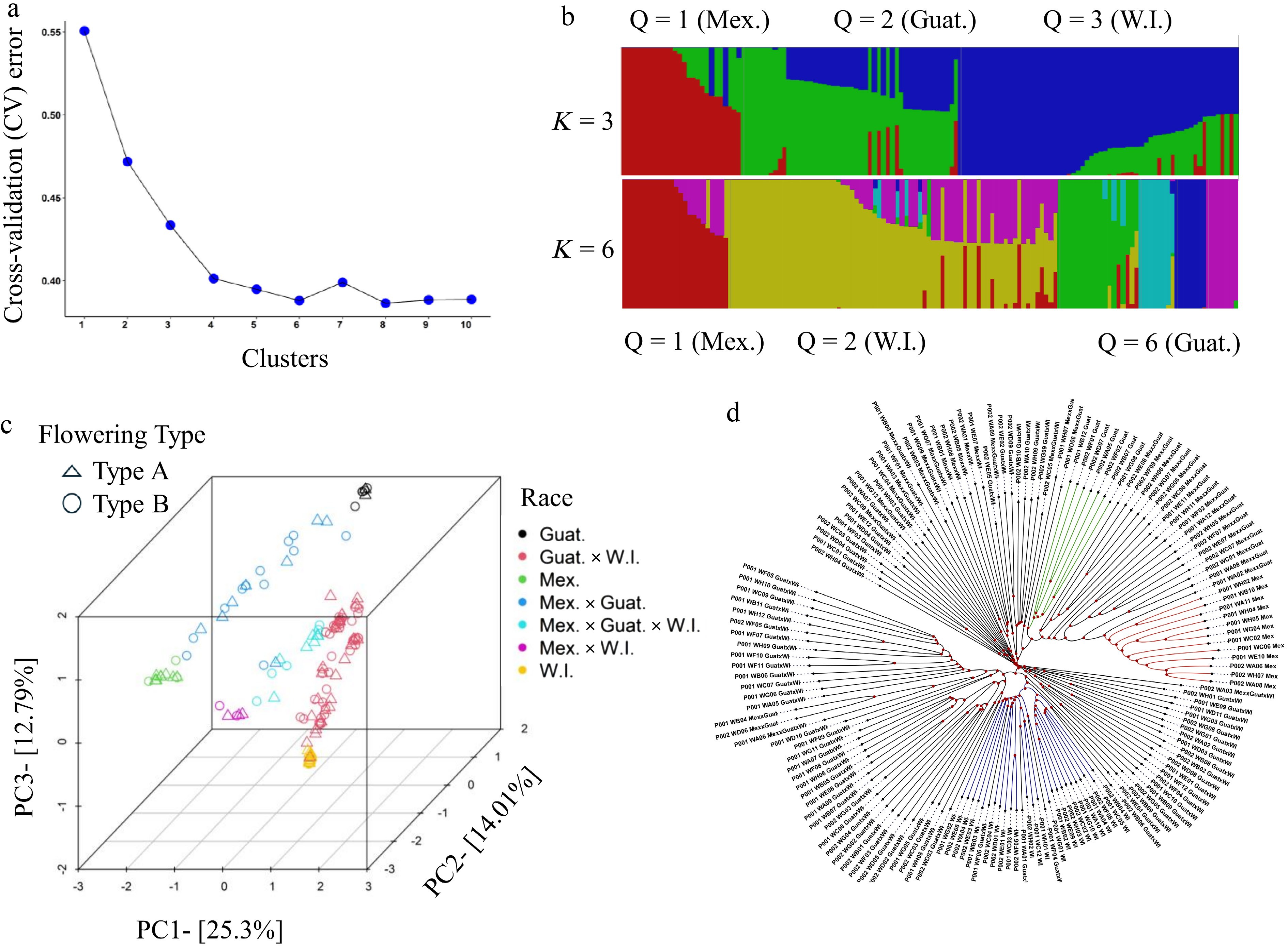
Figure 1.
Population structure analysis of 153 Persea americana accessions. (a) Cross-validation error analysis for clustering with hypothetical K-values ranging from 1 to 10. The x-axis represents K-values (1–10), while the y-axis represents cross-validation error values. The rate of decrease slows after K = 4. (b) Admixture analysis showing population structure for assumed group numbers K = 3 and 6. Each accession is represented as a thin vertical segment, with colors indicating population proportions. Each subgroup is designated by a unique color. In K = 3, accessions with red, green, and blue represent Mexican, Guatemalan, and West Indian races, respectively, and combinations of colors represent hybrid lines. (c) Scatter plot of the first three principal components (PC1, PC2, and PC3). Each dot represents an accession, with symbol shapes indicating flowering types and colors representing different races. (d) Dendrogram of 153 avocado accessions based on whole-genome SNP data (1,072,934 SNPs). Accessions with red, green, and blue represent Mexican, Guatemalan, and West Indian races, respectively. Each accession label is followed by its corresponding race, and the labels can be matched with the IDs and cultivar names listed in Supplementary Table S2.
-
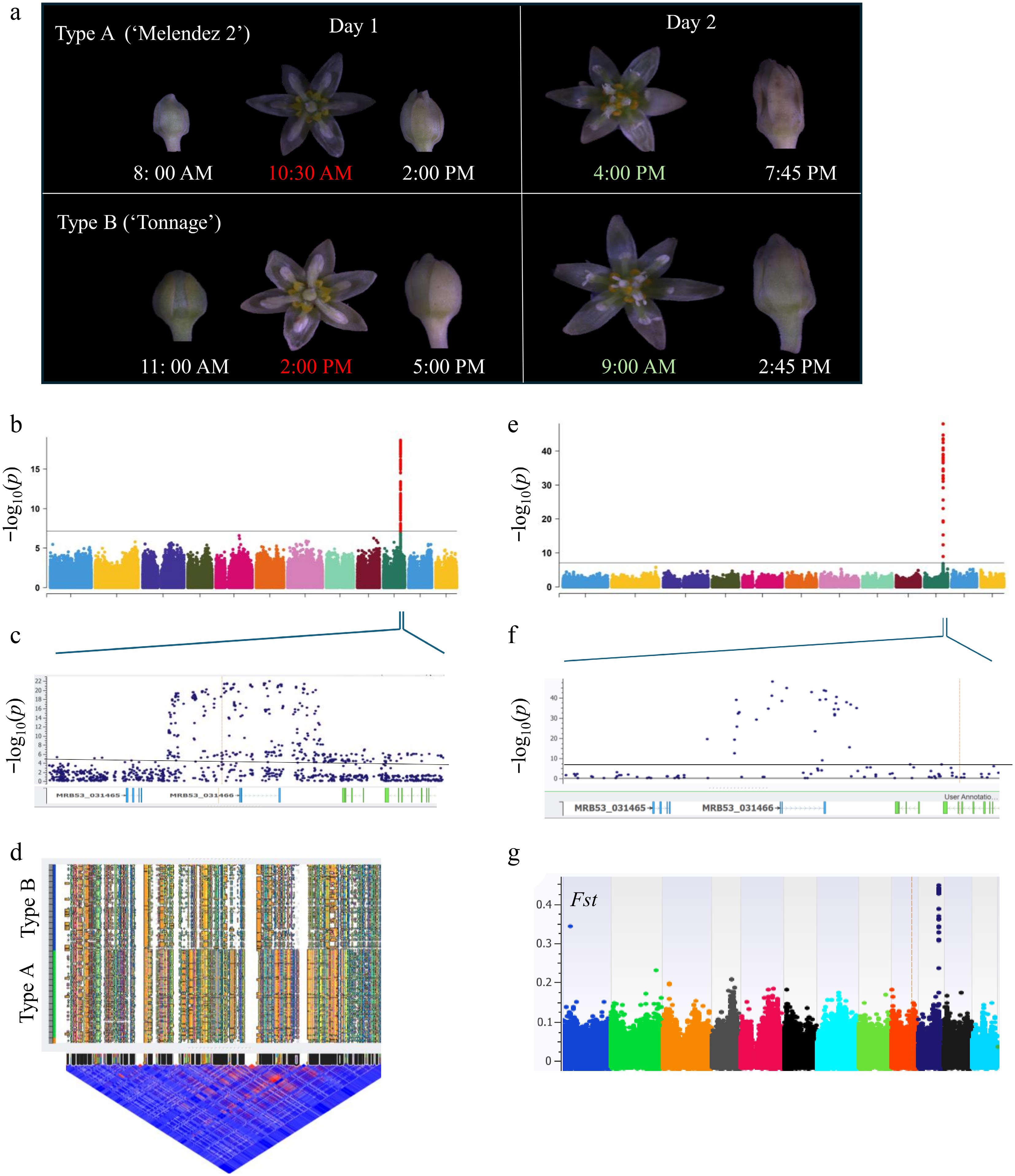
Figure 2.
Genome-wide association study (GWAS) of the flowering type in avocado germplasm using SNP and SV markers. (a) Floral development of Type A ('Melendez 2') and Type B ('Tonnage') from bud to full bloom till flower closing, observed at different times of the day. The photographs illustrate the distinct blooming patterns of both varieties. Both Type A and Type B open as functionally female on the first day with different timing (highlighted in red), followed by functionally male stages on the second day (highlighted in light green). After pollination, the sepals get brown, indicating the end of the pollination period. (b), (e) Manhattan plots illustrating GWAS results for flowering type using (b) 735,030 SNPs and (e) 553,332 structural variants (SV) using the EMAXX model; associations were verified using additional GWAS models (Supplementary Fig. S3). The y-axis represents −log10(p) values. The horizontal line represents 5% Bonferroni correction significance threshold. (c), (f) Zoomed-in genomic regions representing the peak in panels B and E with gene models displayed below the graph. Blue dots represent markers. All significant markers are primarily localized around the MRB53_031466 gene encoding R2R3-type Myb-like transcription factor. (d) Haplotype blocks and Linkage disequilibrium (LD) plots of the flowering type locus. The upper part of the figure illustrates different haplotype blocks in Type A and Type B. The lower part shows an LD heat map of SNPs with red regions representing high LD between adjacent SNPs, while blue regions represent low LD. (g) Fixation Index (FST) plot showcasing genetic differentiation between Type A and Type B avocado accessions. Elevated Fst values indicate regions under potential positive selection, aligning with GWAS signals.
-
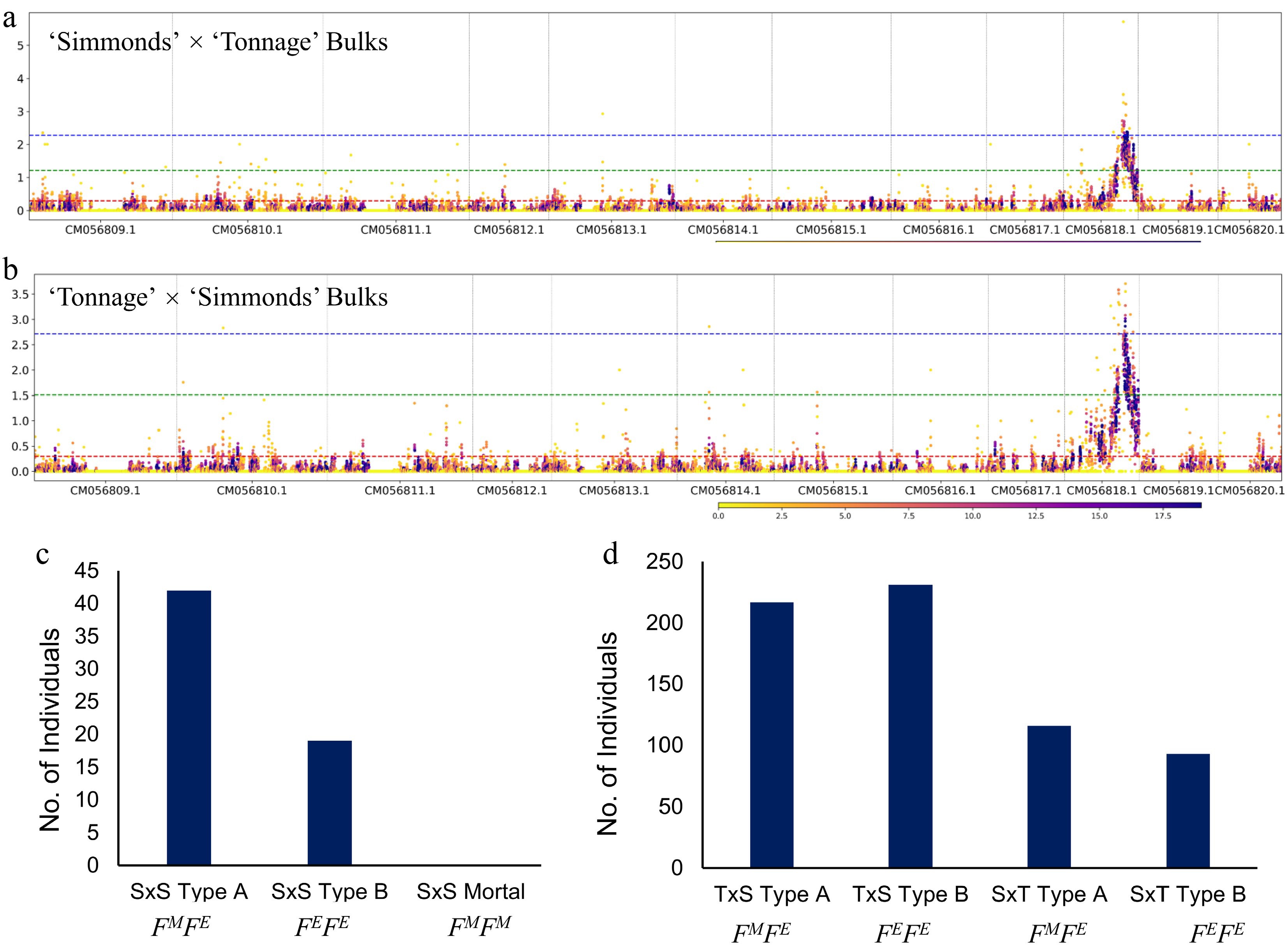
Figure 3.
Bulk segregant analysis maps flowering type to the same loci as GWAS signals on chromosome 10. (a), (b) Bulk segregant analyses (BSA) conducted on both reciprocal avocado populations. (a) 'Simmonds' × 'Tonnage' (n = 50), and (b) 'Tonnage' × 'Simmonds' (n = 50). Segregation ratio of the Type A (FMFE) and Type B (FEFE) genotype in segregating populations: (c) Cv 'Simmonds' selfed, and (d) 'Tonnage' × 'Simmonds' and reciprocal ('Simmonds' × 'Tonnage').
-
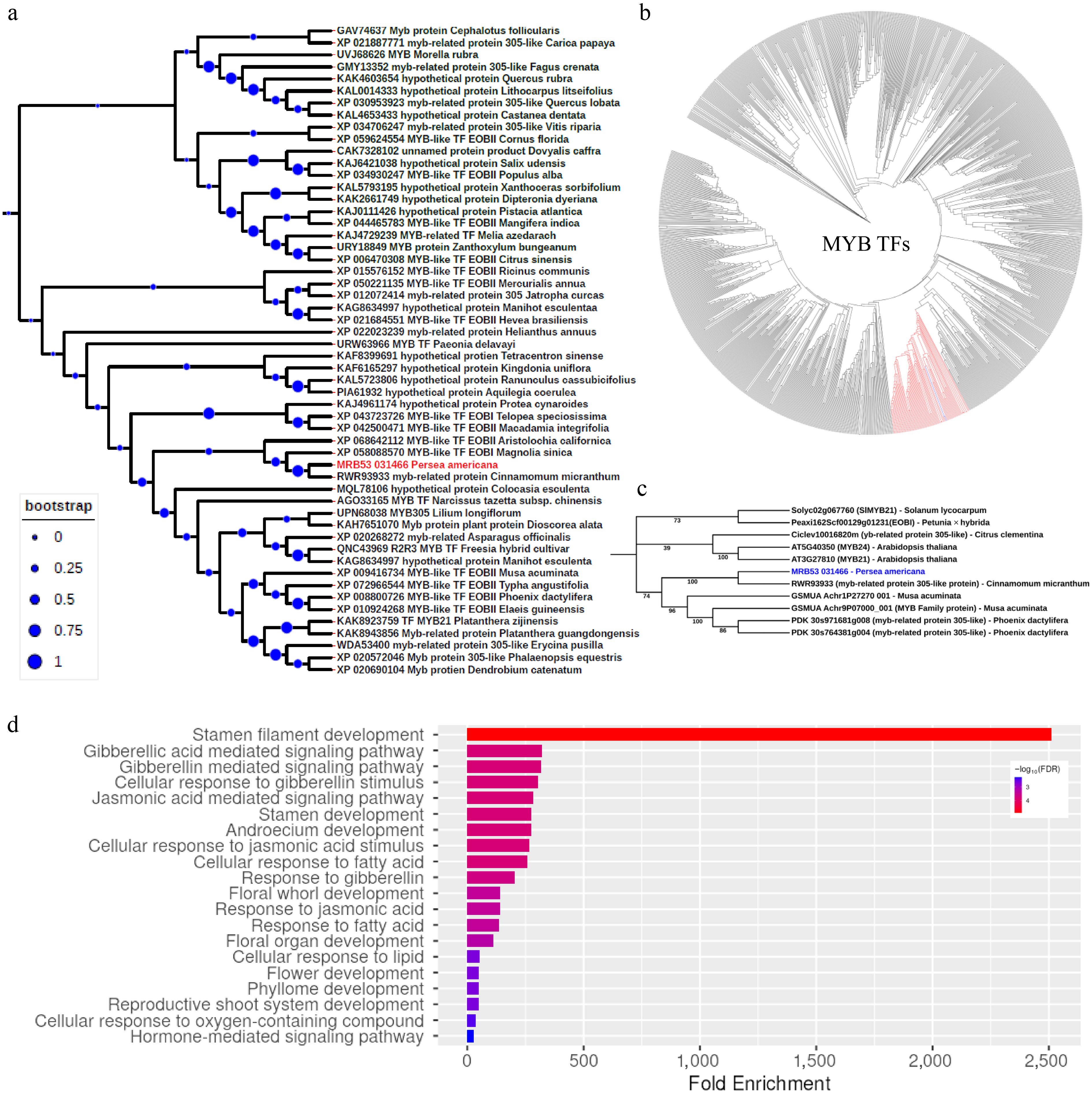
Figure 4.
Phylogenetic analysis of PaDSPD transcription factor. (a) Phylogenetic tree illustrating the evolutionary relationship of PaDSPD proteins from different botanical lineages, revealing it as a homolog of EMMISSION OF BENZOIDS (EOB1), a MYB transcription factor. The evolutionary history was inferred using the Neighbor-Joining method. The bootstrap consensus tree was inferred from 1,000 replicates. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. (b) Phylogenetic tree of MYB transcription factor protein sequences from Arabidopsis thaliana, Solanum lycopersicum, Citrus clementina, Petunia axillaris, Cinnamomum micranthum, Musa acuminata, and Phoenix dactylifera, showing broader evolutionary relationship of EOB1 highlighted in red color. (c) Phylogenetic tree of subgroup 19 within the MYB family, demonstrating the evolutionary placement of PaDSPD highlighted in blue font within this subgroup. (d) Functional enrichment analysis of the Arabidopsis thaliana homologs of PaDSPD, highlighting key biological processes.
-
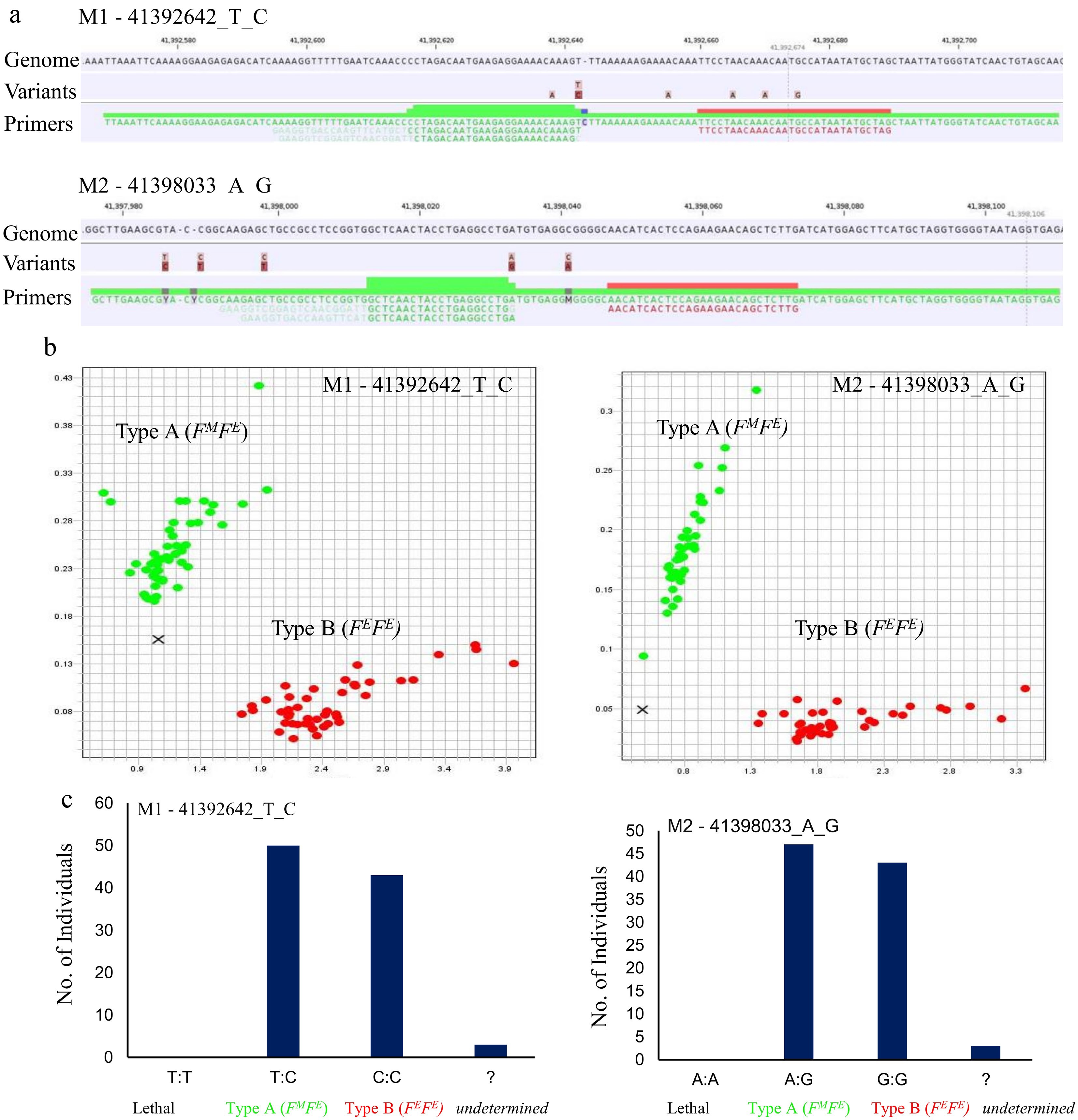
Figure 5.
Development and validation of competitive allele-specific PCR markers for avocado flowering type. (a) Genomic context of the PACE allele-specific primers (FM 'female in the morning' and FE, 'female in the evening' highlighted in green, and the common reverse primer, highlighted in red) targeting two significant SNPs; M1 - 41392642_T_C and M2 - 41398033_A_G. (b) Scatter plots highlighting distinct grouping of accessions into two genetic clusters representing Type A (FMFE) and Type B (FEFE). Red dots represent homozygous alleles and green dots represent heterozygous alleles. X represents undetermined genotypes with ambiguous calls that could not be confidently assigned to either cluster. (c) Bar graphs displaying the calling rates for markers M1 - 41392642_T_C and M2 - 41398033_A_G. The data represent genotype distributions across three categories: Type A (FMFE), Type B (FEFE), and the lethal genotype (FMFM).
-
Observed Expected under a 2:1 expected ratio assuming one homozygotes is lethal Expected under
a 3:1 expected ratioSxS Type A (FMFE)1 42 40.67 45.75 SxS Type B (FEFE) 19 20.33 15.25 Mortal (FMFM) 0 Total 61 χ2 test 0.717 0.268 Table 1.
Segregation of Persea americana cv. 'Simmonds' selfed population for flowering type.
-
Observed Expected
under a 1:1
expected ratioExpected
under a 3:1
expected ratio'Tonnage' × 'Simmonds' Type A (FMFE) 217 224 336 'Tonnage' × 'Simmonds' Type B (FEFE) 231 224 112 Total 448 χ2 test 0.508 1.50865E-38 Reciprocal cross 'Simmonds' × 'Tonnage' Type A (FMFE) 116 104.5 156.75 'Simmonds' × 'Tonnage' Type B (FEFE) 93 104.5 52.25 Total 209 χ2 test 0.112 7.53547E-11 1, Genotypes of Type A and Type B flowering types; FM = 'Female in the morning' allele, and FE = 'Female in the evening' allele of the Persea americana Diurnally Synchronous Protogynous Dichogamy (PaDSPD) gene. Table 2.
Segregation of flowering type in the Florida Avocado Mapping Populations (FLAMP) resulting from a cross between 'Tonnage' × 'Simmonds' and its reciprocal cross 'Simmonds' × 'Tonnage' cultivars.
Figures
(5)
Tables
(2)