HTML
-
Lignocellulose, composed of cellulose, hemicellulose and lignin, is one of the most prevalent and abundant polymers found in the earth's crust (Li et al. 2019). Lignin is bound with carbohydrates (hemicellulose and cellulose) in the plant cell wall, preventing their easy breakdown for bioethanol generation and giving them mechanical resistance (Kataria et al. 2013). From the literature, it has been found that the agricultural and food processing sectors generate a lot of lignocellulose waste every year (Saini et al. 2015). This can be used to make biofuels, food and other products in biorefineries. Lignin protects cellulose and hemicellulose against microbial attack due to its recalcitrant nature. Lignocellulose degradation is an important aspect of carbon recycling in the ecosystem, which is facilitated by the occurrence of lignin degrading microbes (Ragauskas et al. 2006, Bozell & Petersen 2010). Only a few microorganisms can degrade lignin. In particular, bacteria and fungi can depolymerize and mineralize lignin (Janusz et al. 2017).
Ligninolytic enzymes (Lac, MnP and Lip.) are crucial in the environmental breakdown of lignocellulose waste. They are versatile, ubiquitous and renowned for degrading various complexes and recalcitrant polymers (Wong 2009, Chandra & Chowdhary 2015). The activity of these enzymes depends upon the number of parameters (Kumar & Chandra 2020). Due to their multiple applications, ligninolytic enzymes have recently gained extensive attention in biotechnology and related areas (Shi et al. 2013). The biotechnological applications of these enzymes are primarily focused on selecting enzyme producers and yields (Arora & Sharma 2010). Laccases, in particular, are usually employed in the food industry, including beer, wine and fruit juice, as well as baking and the sugar beet pectin gelation processes. It has also been shown to improve food product quality and efficiency (Mayolo-Deloisa et al. 2020). The ligninolytic enzymes have been used in the food industry for decades (Chowdhary et al. 2019). As per the demand of these enzymes in the present era, fungi could act as an efficient source of these enzymes.
Fungi have been observed to grow and multiply in various environments where nutrient sources such as carbon, nitrogen and other elements are accessible (Gupta et al. 2007). As a result, unless the substrate provides all of the nutrients, studying the nutrition of particular fungi is important. Endophytic fungi colonize the intracellular and intercellular parts of plants body without triggering any noticeable disease symptoms (Schulz et al. 2002). They are important sources of industrially important enzymes and bioactive metabolites (Subbulakshmi et al. 2012, Kumar & Prasher 2021). These fungi are also an important source of ligninolytic enzymes. Fungal endophytes can also be used as wood decomposers (Fukasawa et al. 2005). Some fungal endophytes remain inactive till the plant or one of its parts dies, at which point they begin to multiply and become primary colonists (Promputtha et al. 2010). As a result, in the early stages of decomposition, these fungi compete favorably with other saprophytes (Fukasawa et al. 2005). In the literature, there is relatively little information about the potential of endophytic fungi to break down lignin.
In the present study, the effect of different carbon and nitrogen on the growth and ligninolytic enzyme production was analyzed as there was no available information on this fungus in this aspect.
-
The reagents and media used in the experimentation like potato dextrose agar (PDA), yeast extract, agar, fungal DNA isolation kit, sodium hypochlorite (NaOCl), ethanol, chloramphenicol, carboxy-methylcellulose (CMC), sodium chloride (NaCl), congo red, azure B, glucose, peptone, potassium dihydrogen phosphate (KH2PO4), manganese sulfate monohydrate (MnSO4·H2O), magnesium sulfate heptahydrate (MgSO4.7H2O), ammonium tartrate (C4H12N2O6), Ferric sulfate Fe2(SO4)3, Copper sulfate pentahydrate (CuSO4.5H2O), guaiacol, veratryl, fructose, lactose, maltose, pectin, raffinose, sorbose, starch, xylose, sucrose, ammonium acetate, ammonium chloride, ammonium nitrate, ammonium oxalate, ammonium phosphate, ammonium sulphate, ammonium nitrate, sodium nitrate, sodium nitrite and amino acids kit were purchased from the Jaiswal chemical store Chandigarh, India.
-
Asymptomatic leaves, fruits and stems were collected from a healthy mature Dillenia indica L. tree growing at Panjab University, Chandigarh, India. Collected samples were stored in polythene bags and processed within 24 hrs. The samples were first washed with tap water for 30 minutes and then rinsed with double distilled water to remove epiphytes and other debris adhering to the surface. The leaves, stems and fruits were dissected into little pieces of roughly 0.5–1.0 cm. The samples were sterilized by immersing in 80% ethanol for 2 minutes and sodium hypochlorite (NaOCl) solution (3%) for 2–3 minutes, afterward 90% ethanol for the next 2 minutes. The leaf, fruit and stem tissues were rinsed in sterile distilled water and allowed to dry in sterilized conditions (Hallmann et al. (2006). Sterilized samples were then placed in Petri dishes containing potato dextrose agar (PDA) medium supplemented with chloramphenicol (100 mg/l) and incubated for 15–16 days at 24℃. The culture was deposited in culture collection of the Panjab University herbarium (PAN). Faces of fungi number was registered as per Jayasiri et al. (2015).
-
The preliminary identification of an isolated fungus was based on morphological and microscopic characteristics. Morphological charctersitcs such as colony colour, growth rate, texture, shape etc were observed by growing the isolate on PDA plates. Microscopic observations were carried out by preparing slides and stained with cotton blue and observed under the microscope (Matrix VRS-2F) for the nature of hyphae, conidia, conidiophores etc., under 40X- 100X magnifications.
Molecular Identifications
-
The fungal isolate was first grown in PDA plates for 15 days and the mycelia from the fully grown culture were scrapped using a sterilized loop. The DNA was isolated from the mycelia (100 mg) using a fungal DNA extraction Kit. The quantity of DNA was estimated with the help of nanodrops (Thermo ScientificTM NanoDropTM 2000/2000c Spectrophotometers). Using universal primers, the internal transcribed spacer (ITS) rDNA region (ITS1-5.8S-ITS2) was amplified by polymerase chain reaction (PCR). The amplified product was purified and sequenced by Postgraduate Institue of Medical Educatio & Research, (PGIMER) Chandigarh, India. (Sharma et al. 2017). The obtained sequence was searched in NCBI blast to confirm its identity and submitted to the NCBI's GenBank database. After obtaining comparable sequences from the NCBI database, the phylogenetic tree was built using the Neighbor-Joining method with the help of MEGA 7 software.
Preliminary screening of extracellular Cellulase
-
The cellulase screening was performed on glucose yeast extract peptone agar medium supplemented with 1% Carboxy-methylcellulose. The medium was prepared, autoclaved and transferred in petri plates. The petri plates were inoculated with a fungal disc (8 mm) and incubated at 24℃. After 5–7 days, the plates were flooded with 2% Congo red solution and destained for 15 minutes with the help of 1M NaCl. The appearance of clear areas around the fungal colony indicates cellulase activity (Sunitha et al. 2013).
Preliminary screening of Lignin modifying enzyme (LME)
-
Azure-B agar clearance method was used to detect the activity of lignin modifying enzyme. The LME medium (1 liter) was prepared by mixing 0.01 g yeast extract, 1 g KH2PO4, 0.001 g MnSO4·H2O, 0.5 g MgSO4.7H2O, 0.5 g C4H12N2O6, 0.001 g- Fe2(SO4)3, 0.001 g-CuSO4.5H2O. 0.01% Azure B, 1 ml of 20% aqueous glucose solution and 2% agar. The media was autoclaved and transferred to the Petri plates in an aseptic condition. Plates were inoculated with the test fungi and incubated for 7 days at 24℃. After 7 days, discoloration of the blue colour medium was recorded, indicating lignin modifying enzymes activity (Archibald 1992).
Preparation of inoculum and medium
-
The media for the culture was prepared by using 100 ml flasks containing 25 ml of glucose peptone medium (10 g glucose, 2 g peptone, 1.0 g KH2PO4, 0.5 g and MgSO4.7H2O per liter). After that, the medium was autoclaved for 15 minutes at 121 ℃. After cooling, one disc of the test fungus was cut from the margin of a 7 days old culture by using a sterilized cork borer. The flask were inoculated with one fungal disc (8 mm) and incubated for 18 days at 28℃.
Experimentation for nutritional requirements
-
The nutrient requirements for the growth and the ligninolytic enzyme production by the selected fungus were studied by growing in glucose peptone broth at 28℃ with pH 6 and incubated for 18 days. The effect of different carbon sources on the growth and the ligninolytic enzyme production was investigated by replacing the original carbon in the glucose peptone medium with the equivalent amount of different carbon sources. Similarly, in the case of nitrogen, the original nitrogen of the basal medium was replaced with an equivalent amount of different nitrogenous compounds. After 18 days of incubation, the culture was harvested and filtrate and mycelium were separated by filtering it using a pre-weighed Whatman filter paper no. 1. The mycelium was dried in an oven at 60℃ and the mycelium dry weight was measured. The filtrate was centrifuged and used for the quantitative estimation of different ligninolytic enzymes.
Morphological and microscopic identification:
-
Lignin peroxidase (LiP) activity was estimated spectrophotometrically and was based on the oxidation of veratryl alcohol (VA) to veratraldehyde in the presence of H2O2 (Tien & Kirk 1988). The assay solution was comprised of 10 mM VA (0.5 ml) in 0.25 M tartrate acid (0.5 ml) and an aliquot of culture filtrate (0.5 ml). The reaction was started by the addition of 5 mM H2O2 to the solution mixture. The resultant reaction mixture was incubated for 3 minutes at room temperature. The change in absorbance was determined at 310 nm and calculations were made using the molar extinction coefficient (= 9300 M/cm).
Manganese Peroxidase (MnP) activity
-
In order to determine manganese peroxidase activity, Bonnen et al. (1994) methodology was used. The spectrophotometric measurement of guaiacol oxidation in the presence of Mn2+ was used to monitor MnP activity. 0.5 ml of 10 mM citrate buffer (pH 5.5), 0.5 ml of 1 mM MnSO4, 0.4 ml of 1 mM guaiacol, and an aliquot of fungal culture filtrate made up the reaction mixture volume to 2 ml. The reaction was started by adding 0.2 ml of 50 M H2O2 to the reaction mixture. The colour change was recorded in a spectrophotometer at 460 nm after 10 minutes of incubation at room temperature. MnP activity is measured in units of enzyme (IU), with one IU equalling the amount of enzyme that oxidized one micromole of substrate per minute.
Laccase activity
-
Laccase enzyme activity was evaluated using 10 mM guaiacol (0.4 ml) in a 100 mM (1.2 ml) acetate buffer containing 10% acetone (v/) at 30℃. By using a spectrophotometer, the reaction mixture with colour change was recorded at 470 nm after 5 minutes of incubation. The Laccase enzyme activity was estimated using an extinction coefficient of 6740 M/cm and represented as International Units (IU) (1 mol of substrate conversion/min) (Das et al. 1997).
Statistical analysis
-
All the experiments were performed in triplicates. By using Statistical Package for the Social Sciences (SPSS) 16 software, the recorded data was analyzed using One-way Analysis of Variance (ANOVA), and Tukey's test was used to evaluate significance (P = 0.05). The data has been given as a mean with a standard deviation (S.E.).
Lignin Peroxidase (LiP) activity
-
The morphological observations revealed that the colony was initially white with cottony aerial mycelium, having fringed margin, turned dark brown later on. Pycnidia were usually single, globose to subglobose, unilocular, dark brown or black in colour. Conidiogenous cells were hyaline, cylindrical, tapering towards the apex, 8–13× 1.5–2 μm. Conidia were unicellular, hyaline, ellipsoidal to cylindrical, rounded at both ends, 5–8 × 1.8–2 μm (Fig. 1). Based on these morphological and microscopic observations, the isolate was identified as Diaporthe sp. For further confirmations, DNA was isolated from the fully-grown culture and ITS 5.8S rDNA region was amplified, purified and sequenced. ITS 5.8S rDNA analysis followed by search in NCBI blast search tool showed that the fungus was closely related to Diaporthe phaseolorum (Fig. 2). Morphological, microscopic and molecular analysis confirmed the identification as Diaporthe phaseolorum. The obtained ITS sequence was submitted to GenBank for accession number (MK757169.1). The taxonomic description along with other details was also submitted to the Faces of Fungi database (FoF 10638).
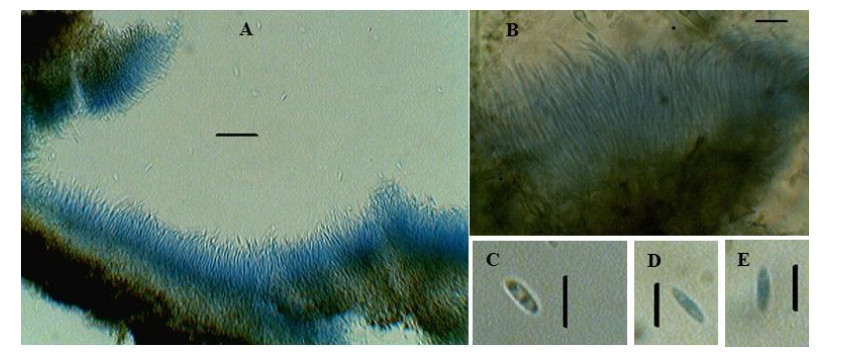
Figure 1. Diaporthe phaseolorum. A, B Conidiophore bearing conidia. C, D, E Conidia. Scale Bars: A–B = 20 μm, C–D = 10 μm.
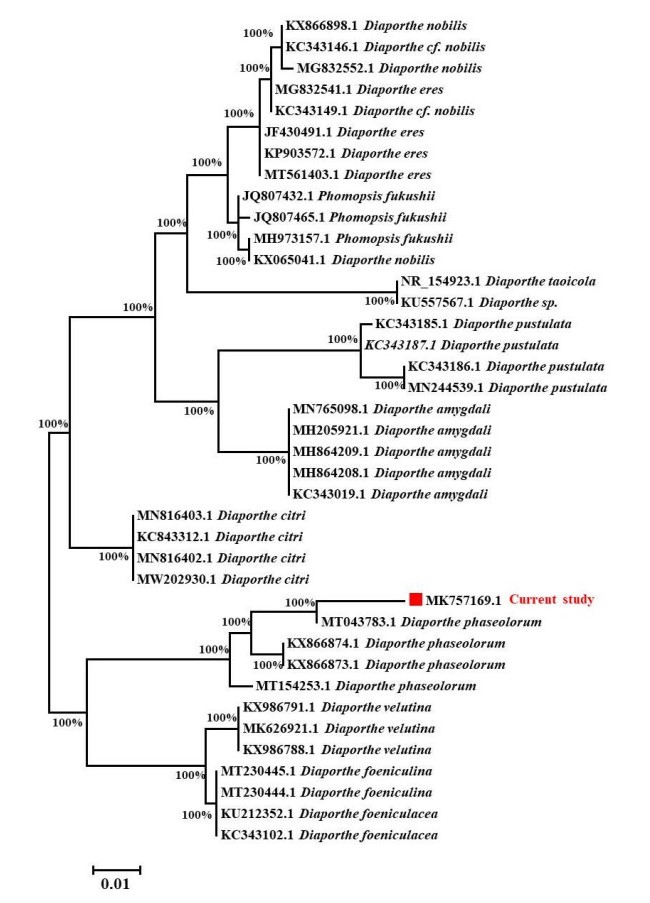
Figure 2. Phytogenic analysis of isolated fungus with its related species by comparing ITS gene sequences obtained from NCBI GenBank. The evolutionary history was deduced using the TamuraNei model and the Neighbor-Joining method with 1000 replications bootstrap. It is shown the Tree shows highest log likelihood (-1382.58). The branch lengths are measured in the number of substitutions per site, and the tree is depicted to scale. A total of 39 nucleotide sequences were examined. 1st+2nd+3rd+Noncoding codon locations were included. Gaps and missing data were removed from all positions. MEGA7 was used to conduct evolutionary analysis with bootstrap values higher than 50% is given in each branch.
Extracellular cellulase and ligninolytic activity
-
When the selected fungal species was screened for lignocellulolytic enzymes using the CMC and LME assays, the radial mycelial growth and decolourization area having 2 distinct colour removal patterns were detected. The formation of a clear zone around the colonies showed cellulase activity. The presence of lignin and manganese peroxidase was confirmed by the absence of blue colour around the colonies on the assay plates (Fig. 3).
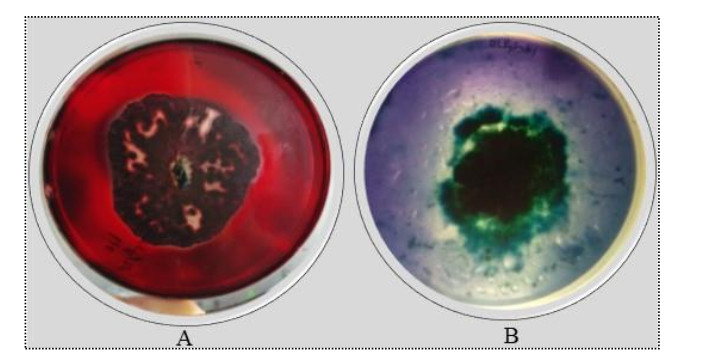
Figure 3. Qualitative screening of Diapothe phaseolorum for cellulolytic and ligninolytic enzymes. A cellulase activity. B ligninolytic enzymes activity.
Effect of different Carbon's on growth and ligninolytic enzyme production
-
Diaporthe phaseolorum showed a different pattern of growth in different carbon sources. Most of the carbon sources examined showed moderate growth, but sucrose showed the highest growth in comparison to other carbons, as shown in Fig. 4. Medium without carbon sources showed very little growth. The carbon sources along with dry mycelium weight in their decreasing order were: sucrose > glucose > pectin > lactose > raffinose > maltose > sorbose > starch > fructose > xylose > control (Fig. 4). As a result, Diaporthe phaseolorum can be cultured utilizing any of the mentioned carbon sources. Different carbon sources had a significant impact on laccase activity. Pectin had the highest laccase activity i.e., 44.5 U/ml, followed by starch and glucose. No enzyme activity was observed in the control medium. In xylose and control, laccase activity was very low. LiP activity was highest in fructose, followed by pectin and glucose. In medium containing fructose, LiP activity was 3.5 U/ml. Similarly, in the case of MnP, the medium containing glucose showed the highest MnP production, followed by starch, sucrose and raffinose. The MnP activity was 6.88 U/ml in medium supplemented with glucose. There was no MnP activity in the medium without a carbon source. In the absence of a carbon source, there was no enzymatic activity. Among three enzymes, laccase activity was found maximum. The growth of Dipothe phaseolorum and ligninolytic enzymes production in different carbon sources are given below in Fig. 4.
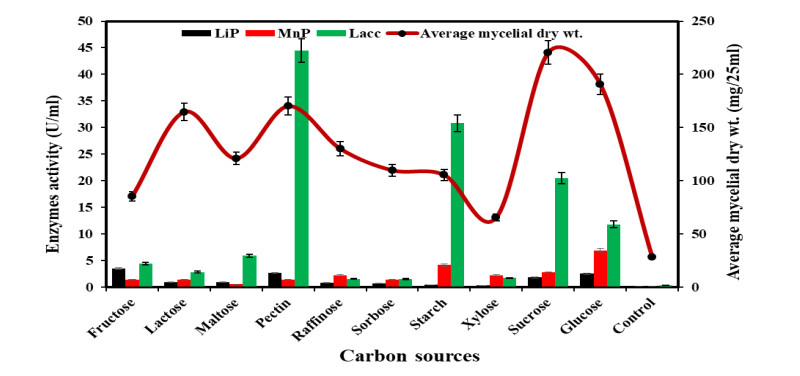
Figure 4. Growth and ligninolytic enzyme production by Diaporthe phaseolorum in different carbon compounds.
Effect of different Inorganic nitrogen on growth and ligninolytic enzyme production
-
Among all of the studied inorganic nitrogen sources, potassium nitrate and sodium nitrate were the best inorganic nitrogen sources for the growth of Diaporthe phaseolorum. The laccase enzyme production was highest in a medium containing potassium nitrate followed by sodium nitrate and ammonium sulphate. The laccase activity was 62.9 U/ml in a medium containing potassium nitrate. The LiP and MnP activities were maximum in medium supplemented with ammonium acetate and ammonium phosphate; and were about 20.6 U/ml and 19.2 U/ml. Fig. 5 depicts the growth of Diapothe phaseolorum and the production of ligninolytic enzymes in various inorganic nitrogen sources.
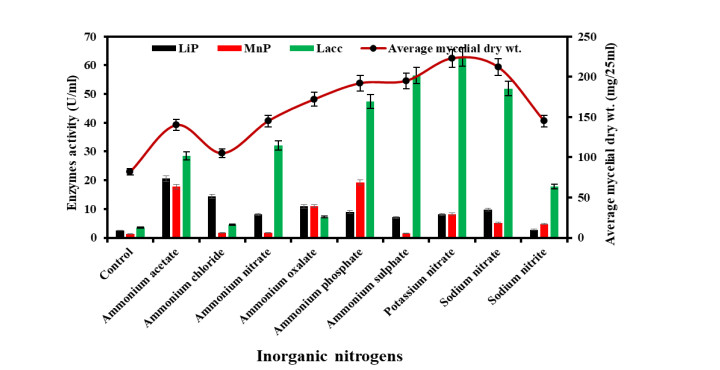
Figure 5. Growth and ligninolytic enzyme production by Diaporthe phaseolorum in different inorganic nitrogen compounds.
Effect of organic nitrogen on growth and ligninolytic enzyme production
-
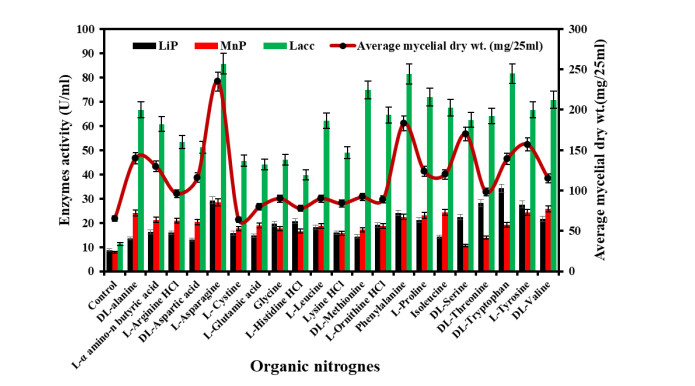
The medium enriched with DL-asparagine was the best for mycelia growth among twentyone different organic nitrogen sources. There was limited growth in control, cystine and L-histidine HCL supplemented medium. In decreasing order, mycelial growth in relation to amino acids: Lasparagine > phenylalanine > DL-alanine > serine > tyrosine > DL – alanine > DL-tryptophan > L- α amino-n butyric acid > L-proline > isoleucine > DL-aspartic acid > DL valine > DL-threonine > L-arginine HCL > DL-methionine > glycine = L-leucine > lysine HCL > L-histidine HCL > Lcystine > control. DL-tryptophan acts as an inducer in LiP production, the maximum production of LiP was observed in a medium containing DL-tryptophan (34.2 U/ml) followed by L-asparagine, DL-threonine, L- tyrosine and phenylalanine. MnP activity was highest in medium supplemented with L-tyrosine followed by DL-valine, L-asparagine, isoleucine and DL-alanine. Similarly, the laccase production was maximum in L-asparagine, followed by DL-tryptophan and phenylalanine. The growth of Diapothe phaseolorum along with ligninolytic enzymes production in different organic nitrogen sources are given below in Fig. 6.
Identification
-
Endophytes are the most important sources of industrially important enzymes having broad application (Fouda et al. 2015). Endophytic strains of the family Xylariaceae have been found to have ligninolytic activity in agar medium using Poly R-478 as an indicator (Urairuj et al. 2003). The physiology and metabolism of enzyme synthesis by fungi require carbon and nitrogen nutrients (Kanwal & Reddy 2011). This study studied the effects of different carbon and nitrogen sources on the growth and ligninolytic enzyme production by the endophytic fungus Diaporthe phaseolorum. Different fungal strains require different nutrient sources for their growth, enzyme production and other metabolic processes because all fungi cannot grow on the same substrate. For fungi to grow, carbon is the most crucial component and accounts for around half of the total dry weight of the fungus (Zhang & Elser 2017)
In the present study, Sucrose was found to be the best carbon source for the growth of Diaporthe phaseolorum. Xylose was the optimal medium for the growth of Chondrostereum purpureum (Prasher & Shehnaz 2019) and Flagellospora penicillioides (Sati & Bisht 2006). In previous studies, glucose appeared to be the most effective growth promoter among the available carbon sources, followed by sucrose (Dhanda & Prasher 2021). In respect of fungal growth and development, maltose and fructose are recognized as good sources of carbon (Held 1970, Singh & Tandon 1970). Glucose and sucrose are the simplest carbon sources that can be easily broken down by the fungal strains, hence making them easy to utilize in different metabolic pathways. Hence, they are preferred as an exceptional carbon source for most fungi. It has also been found that the production of ligninolytic enzymes (Laccase, MnP, and LiP) is not influenced only by the fungal type/strain but also by the culture conditions, carbon and nutrient sources (carbon and nitrogen) and their concentrations (Stajic et al. 2006).
The efficacy of ligninolytic enzymes was found to be strongly controlled by carbon and nitrogen sources in the current study. Fructose, glucose, and pectin were shown to be the most effective carbon sources for LiP, MnP and Lacc synthesis compared to the other carbon sources studied. Iqbal et al. (2011) reported that starch and raffinose were the optimal carbon sources for the production of LiP, starch and sucrose for MnP production, and sucrose and maltose for laccase production by Trametes versicolor. Ligninolytic enzyme production is influenced by the amount of carbon in the nutritional medium and the lignocellulosic substrate. Galhaup et al. (2002), reported that Trametes pubescens started producing laccase when the glucose levels in the growth medium reached a particular low, critical level.
Earlier studies have shown that the nature and quantity of nitrogen sources are major nutritional factors in regulating the production of ligninolytic enzymes (Levin et al. 2010). Nitrogen plays an important part in the physiology of fungi and as it makes up about 38% of the dry weight mass (Zhang & Elser 2017). As per studies (Elisashvili et al. 2001, Galhaup et al. 2002), the nature and quantity of nitrogen supplies are significant nutritional factors in the growth and ligninolytic enzymes. In the present study, potassium nitrate, sodium nitrate and ammonium sulphate were the best media for the growth of Diaporthe phaseolorum. In the earlier study, Grammothele fuligo showed optimal mycelial growth in ammonium chloride (Chauhan 2019). The laccase activity was 186.2 U/ml in a medium containing ammonium nitrate. The LiP and MnP activities were maximum in medium supplemented with ammonium acetate and ammonium phosphate and were about 61.82 U/ml and 53.17 U/ml.
Sodium nitrate was reported as the best nitrogenous compound for laccase production by Penicillium martensii (Elshafei et al. 2012). Phanerochaete chrysosporium produces a good amount of LiP and MnP in media containing ammonium chloride (Varshney et al. 2013). Similarly, Kachlishvili et al. (2006), studied the effect of different nitrogen compounds on the production of ligninolytic enzymes by white rot fungi such as Funalia trogii, Lentinus edodes, Pleurotus dryinus, and P. tuberregium. The results revealed that the ligninolytic enzymes production increased when medium supplemented with additional nitrogen compounds. The enzymes production was increased in the medium due to higher biomass in the nitrogen supplemented media.
DL-asparagine gave optimal growth of Diaporthe phaseolorum and LiP, MnP and Laccase production was recorded highest in DL-tryptophan, DL-threonine and DL-methionine. L-tyrosine inhibits the growth of Curvularia senegalensis, Curvularia prasadii and Phoma vulgaris (Dandge 2012). Levin et al. (2010) observed that laccase and MnP production by Coriolus versicolor and Trametes trogii in glutamic acid (nitrogen source). Carbon and nitrogen sources with a greater ability to produce ligninolytic enzymes can be used to produce these enzymes in a commercial scale.
-
The study presents intriguing information about the fungus's growth and production of ligninolytic enzymes. The oxidation potential of the fungus's ligninolytic enzymes indicates that, it can produce these enzymes commercially for a variety of industrial applications. This data can also be used to produce mycelial and ligninolytic enzymes and can also be used on a commercial scale for environmental remediation of industrial contaminants.
-
The authors acknowledge the Department of Botany, Panjab University Chandigarh, India, for providing infrastructure and instrumentation. Vijay Kumar is also thankful for Senior Research Fellowship (File No. 09/135(0854)/2019-EMR-I) by the Council of Scientific and Industrial Research (CSIR), India, during research work.
- Copyright: © 2021 by the author(s). This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
| V Kumar, IB Prasher. 2021. Ligninolytic enzymes production by endophytic fungus Diaporthe phaseolorum (Desm.) Sacc. under the influence of different carbon and nitrogen sources. Studies in Fungi 6(1):531−542 doi: 10.5943/sif/6/1/43 |