-
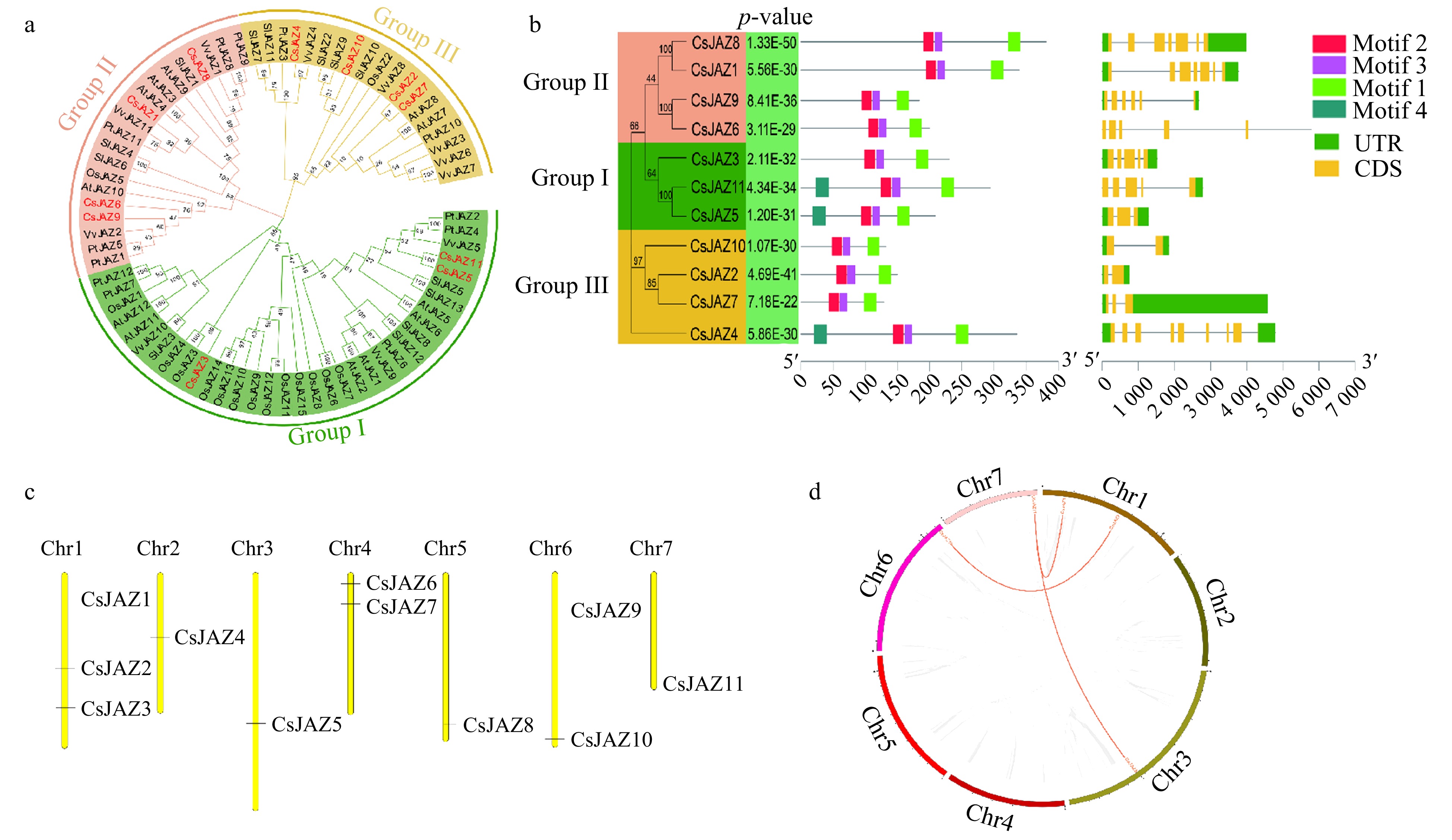
Figure 1.
(a) Phylogenetic analysis of JAZ proteins in 11 C. sativus (Cs), 12 A. thaliana (At), 13 S. lycopersicum (Sl), 15 O. sativa (Os), 11 V. vinifera (Vv), and 12 P. trichocarpa (Pt). ClustalW multi-sequence alignment was performed and an unrooted phylogenetic neighbor-joining tree was constructed fromon JAZ protein by using MEGA X software. Genes IDs are listed in Supplemental Table S3. (b) Phylogeny, gene structure, and motif compositions of the JAZ gene family in cucumber. (c) Chromosome distribution of CsJAZ in the genome of cucumber. The chromosome numbers were demonstrated at the top of each bar and the scale is in megabases (Mb). (d) Gene duplication of CsJAZ genes on 7 chromosomes of cucumber genome. Gray lines represent all syntenic blocks in the cucumber genome. Duplicated gene pairs are identified with red lines inside the circle. Each chromosome number is represented using different colours.
-
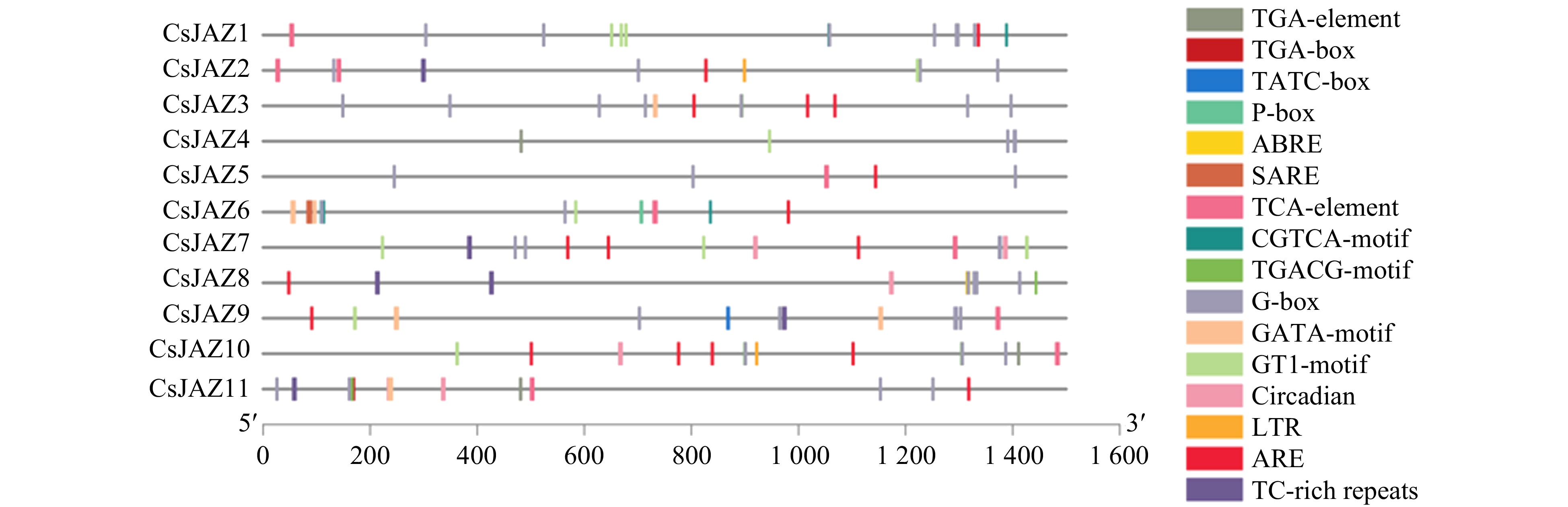
Figure 2.
Prediction of cis-acting elements in CsJAZ promoters associated with plant hormones and stress are shown. The distribution of cis-acting elements in the 1.5 Kbp upstream promoter region of the CsJAZ genes associated with plant hormones, grow and stress are illustrated.
-

Figure 3.
Morphological changes and statistical collation in response to waterlogging and waterlogging+MeJA. (a) The response of WL and WL+MeJA after 3, 5, 7 d waterlogging treatment. (b) The survival rate of WL and WL+MeJA after 7 d of waterlogging treatment. (c) Comparison of the adventitious root (AR) number capacity of WL and WL+MeJA after 7 d of waterlogging treatment. ** p < 0.01.
-
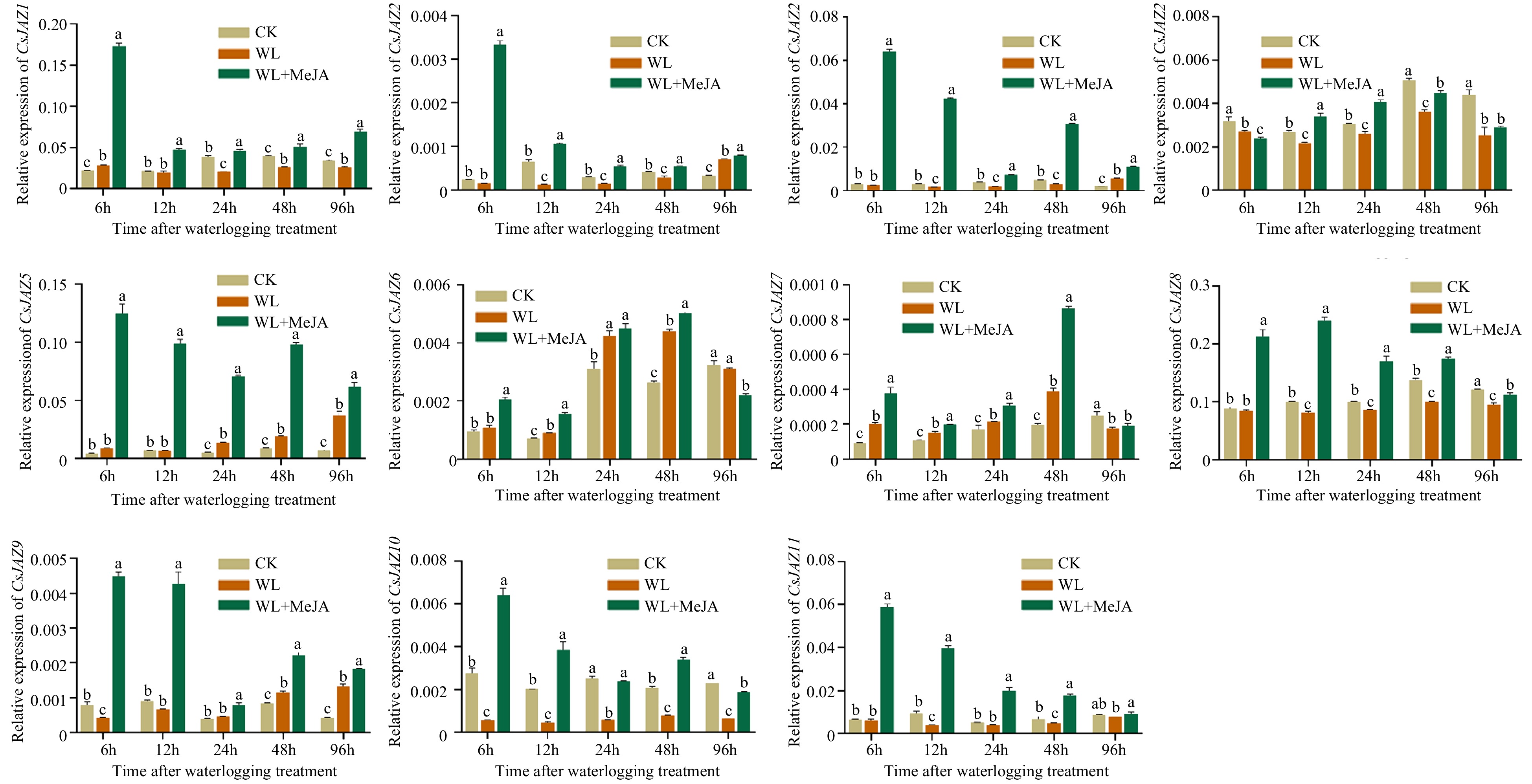
Figure 4.
Expression of CsJAZ genes in response to waterlogging treatment. The different lowercase letters above the bars indicate the significance (p < 0.05) of the relative expression level between two samples. The data for quantitative RT-PCR analysis were the mean and standard deviation of three bio-duplicates in the hypocotyl and three technical duplicates in each biological sample. The Y-axis is the relative level of expression.
-
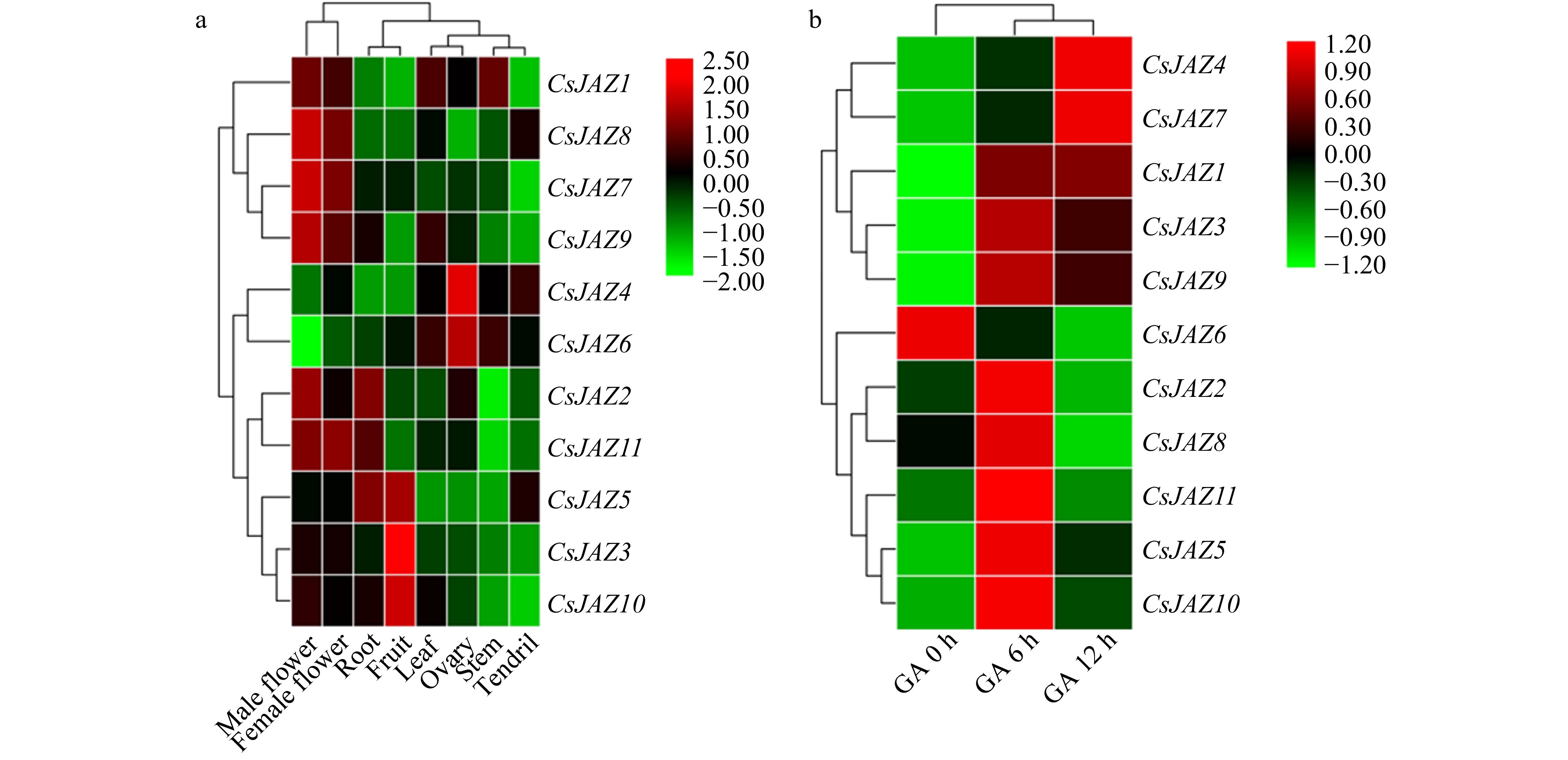
Figure 5.
Heatmap representation of CsJAZ genes in different tissue and post exogenous GA treatment of cucumber. The log2 transformation method was used to normalize and convert the RPKM values and displayed on the colored scale bar.
-

Figure 6.
Subcellular localization analysis of CsJAZ8-GFP fusion protein transiently expressed in Nicotiana tobaccum L. (a) Schematic representation of constructs used for subcellular localization of the CsJAZ8 protein. (b) The photographs were taken using green fluorescence (GFP), visible light, and merged light. Bar = 50 μm.
-
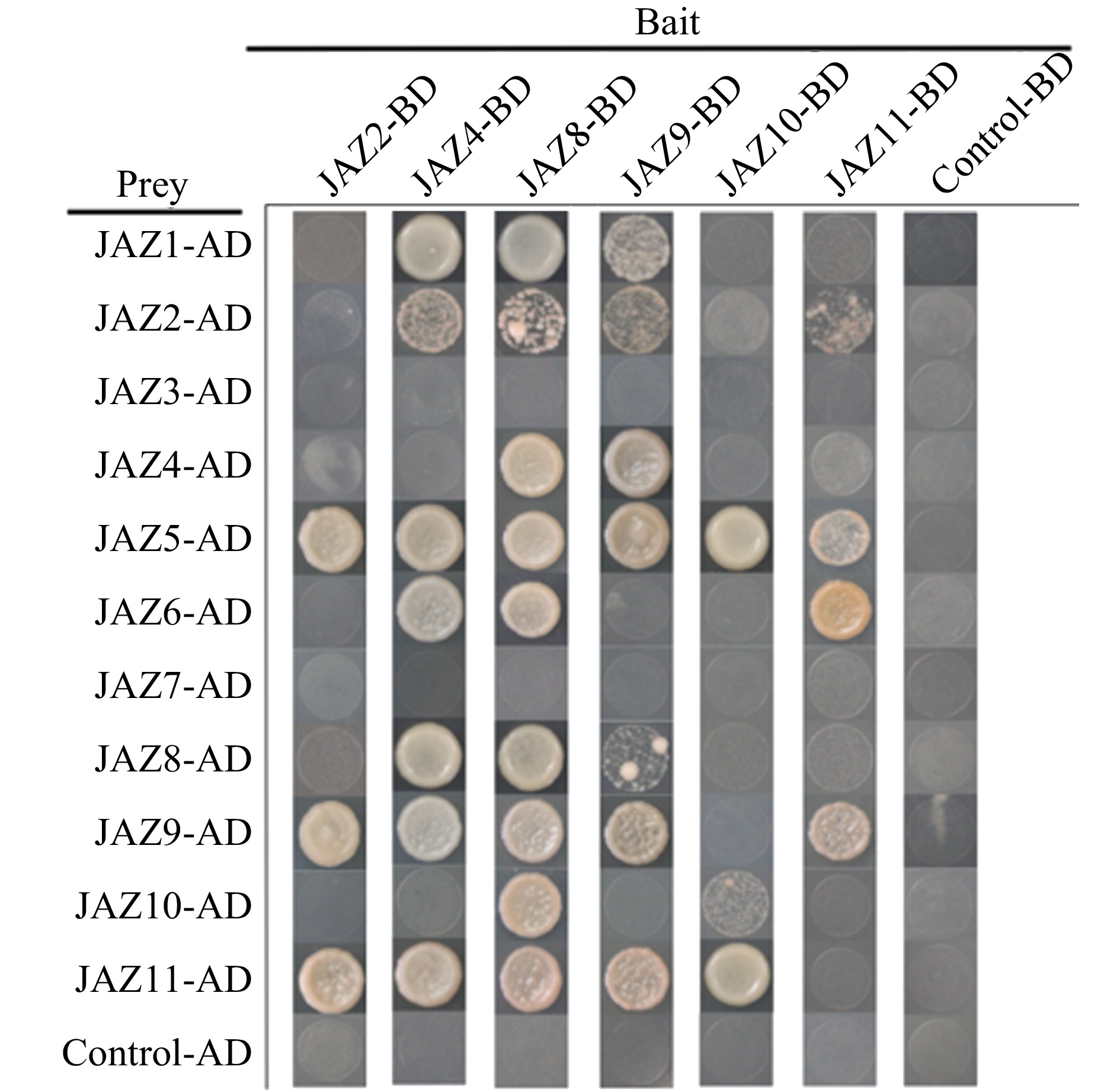
Figure 7.
Yeast two-hybrid analysis of Homo- and heteromeric interactions in CsJAZ proteins. The full-length CsJAZ was fused with pGADT7 and pGBKT7 to generate prey and bait vectors, respectively, and then transformed into the yeast strain AH109. The interactions were determined by growth on SD-Trp/-Leu/-His/-Ade nutritional selection medium. The mating of pGBKT7-53/pGADT7-T7 was used as a positive control, and pGBKT7-Lam/pGADT7-T7 was used as a negative control.
-
Gene name Gene identifier Chr Start (bp) End (bp) ORF (aa) MW (KDa) PI GRAVY CsJAZ1 Csa1G042920 1 4573626 4577405 340 36 619.75 9.2 −0.25 CsJAZ2 Csa1G435720 1 15995968 15996717 150 16 930.92 7.21 −0.767 CsJAZ3 Csa1G597690 1 22692966 22694494 231 25 028.26 9.98 −0.455 CsJAZ4 Csa2G222060 2 10711729 10716520 336 36 967.37 6.71 −0.816 CsJAZ5 Csa3G645940 3 25350295 25351576 209 22 874.76 8.94 −0.631 CsJAZ6 Csa4G009880 4 1470022 1476285 200 22 487.17 6.98 −0.553 CsJAZ7 Csa4G062400 4 4986428 4991013 129 14 861.18 10.42 −0.425 CsJAZ8 Csa5G628650 5 25425307 25429296 381 39 666.79 9.3 −0.236 CsJAZ9 Csa6G091930 6 6249153 6251831 184 20 306.27 9.41 −0.317 CsJAZ10 Csa6G523460 6 28126619 28128465 132 14 743.72 9.5 −0.805 CsJAZ11 Csa7G448810 7 18408423 18411214 295 32 092.09 9.31 −0.758 Table 1.
Summary information on the CsJAZ gene family in cucumber.
-
Motif E-value Sites Width Sequence Logo 1 1.6e-077 11 20 DLPJARKASLQRFLEKRKDR 
2 5.7e-074 11 15 QMTIFYNGKVCVYBD 
3 2.6e-0.13 11 15 PEDKAKAIMALASKG 
4 4.0e-003 3 21 EKSNFAQTCNLLSQYLKEKRT 
Table 2.
Information on conserved motifs in cucumber JAZ proteins.
Figures
(7)
Tables
(2)