-
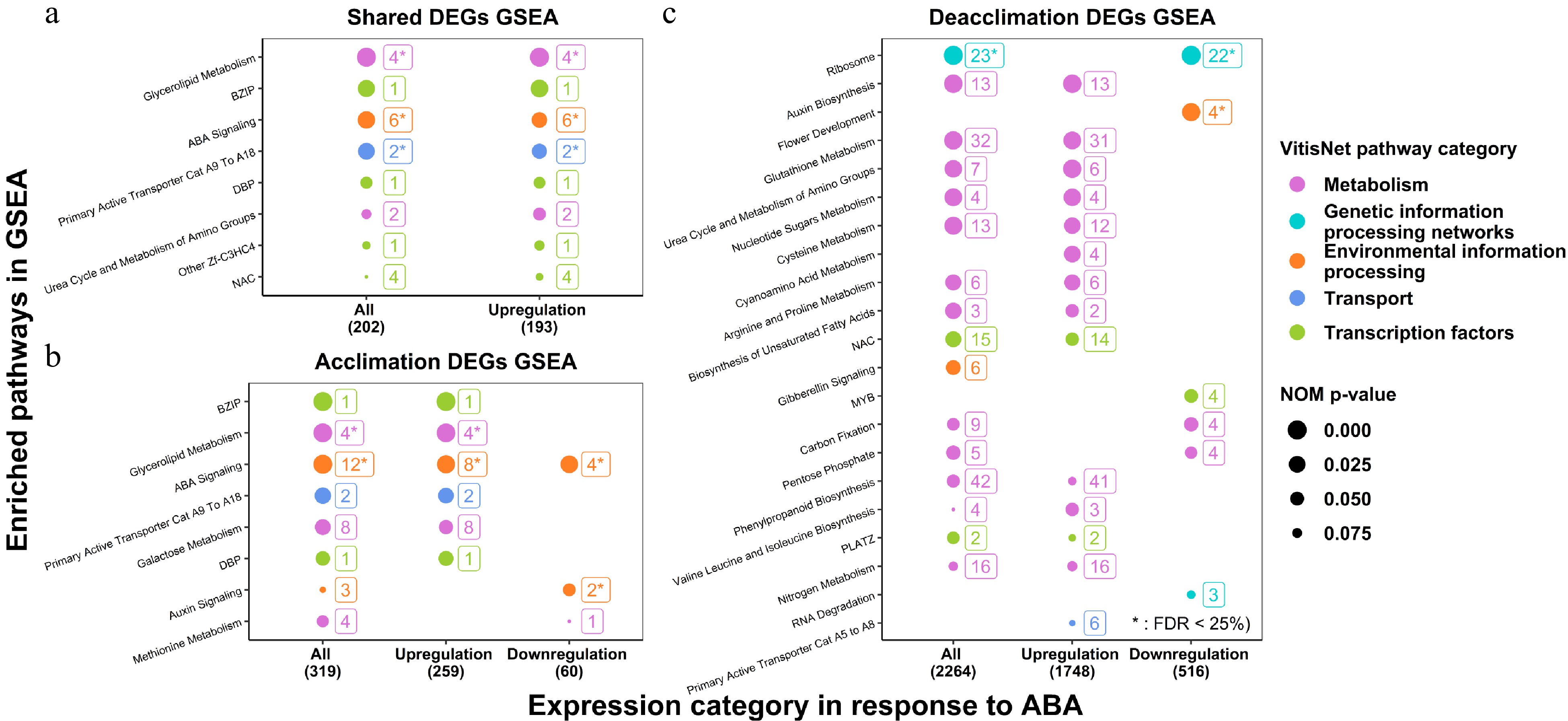
Figure 1. GSEA of ABA-induced DEGs in the acclimation and the deacclimation experiments. (a) GSEA of shared DEGs in two experiments; (b) GSEA of DEGs in the acclimation experiment; (c) GSEA of DEGs in the deacclimation experiment. Only the pathways with NOM p-value < 0.1 are shown.
-
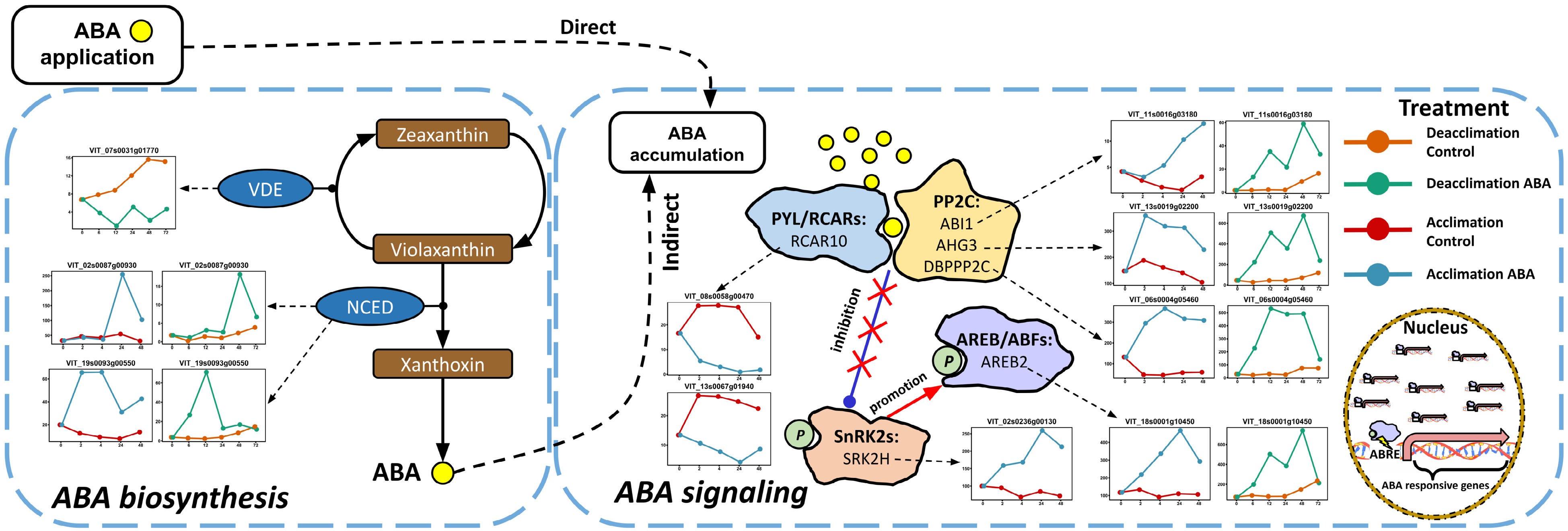
Figure 2. Impact of ABA application on ABA biosynthesis and signaling pathways in the acclimation and the deacclimation experiments. The pathways were reduced based on the full pathways in VitisNet. The expression value of genes was normalized by DESeq2. Normalized expressions of the DEGs observed in the deacclimation experiment for control and ABA treatment are orange and green, respectively. Normalized expressions of the DEGs observed in the acclimation experiment for control and ABA treatment are red and blue respectively. All shared and unshared ABA-induced DEGs are shown. Abbreviations: ABRE, ABA responsive element; AREB/ABF, ABRE-binding protein/ABRE-binding factor; NCED, 9-cis-epoxycarotenoid dioxygenase; PP2C, group A protein phosphatase 2C; PYL/RCAR, pyrabactin resistance 1-like protein/regulatory component of ABA receptor; SnRK2, sucrose non-fermenting 1-related protein kinase 2; VDE, violaxanthin de-epoxidase.
-
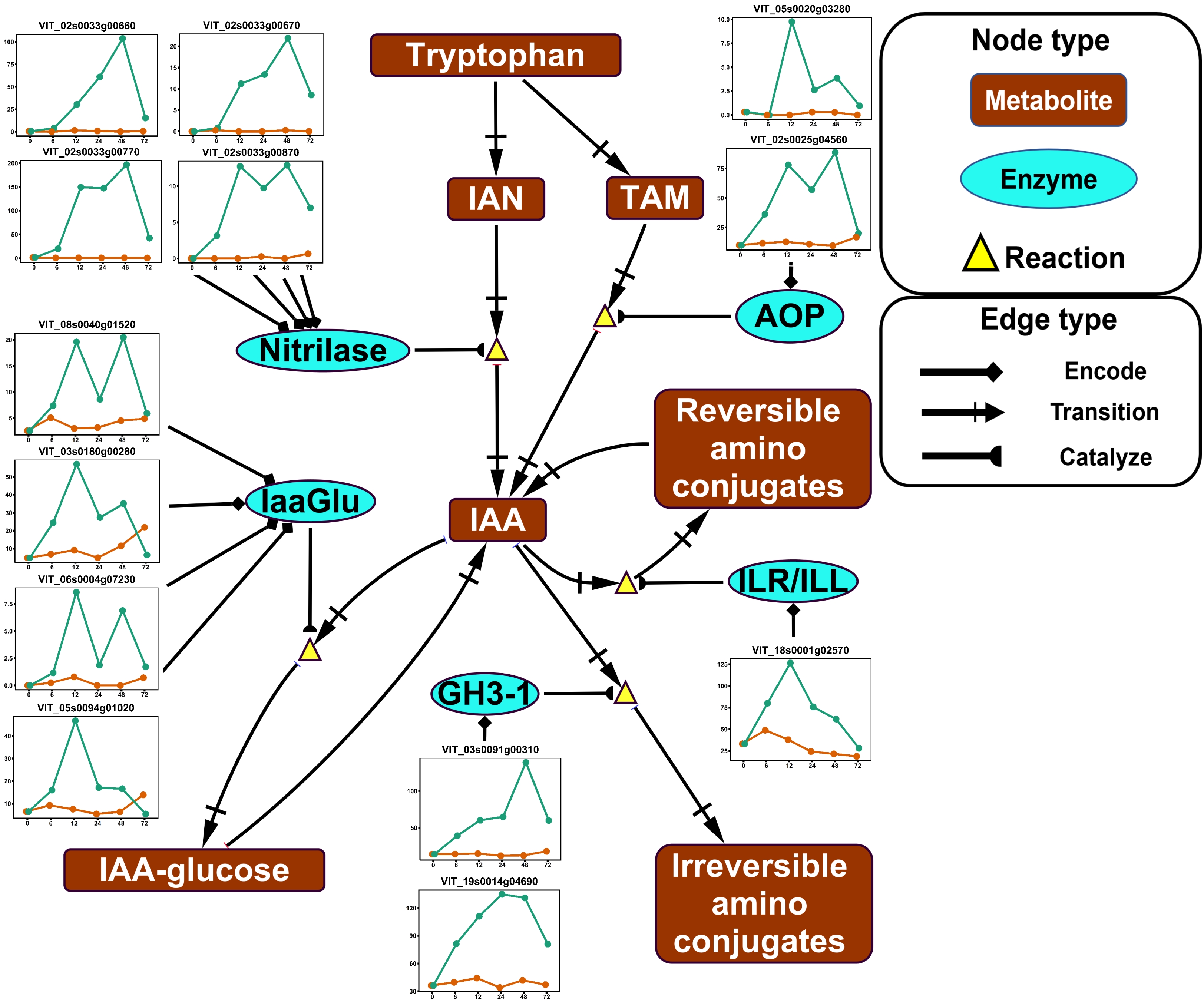
Figure 3. Impact of ABA application on auxin (IAA) metabolism in the deacclimation experiment. The pathway was reduced based on the full pathway in VitisNet. The expression value of genes was normalized by DESeq2. Normalized expressions for control and ABA treatment groups are shown by orange and green lines, respectively. Abbreviations: AOP, amine ocidase flavin containing; GH3, GRETCHEN HAGEN 3; IAA, indoleacetic acid; IaaGlu, indoleacetic acid glucosyltransferase; IAN, indole-3-acetonitrile; ILR/ILL, IAA-Leu resistant/ILR-like; TAM, tryptamine. Refer to Fig. 2 for keys of treatments in gene expression plots.
-
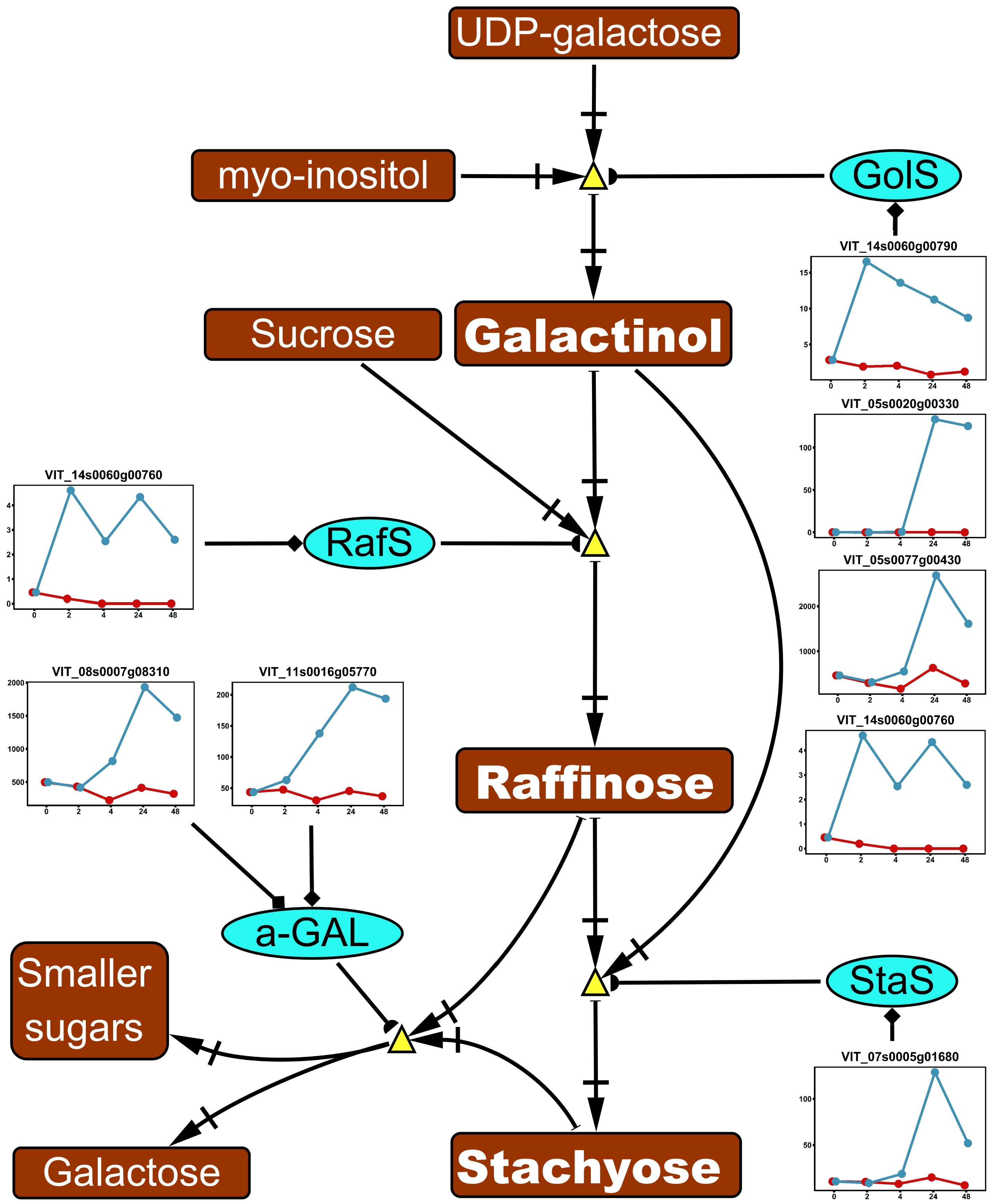
Figure 4. Impact of ABA application on raffinose family oligosacchrides (RFOs) metabolism in the acclimation experiment. The pathway was reduced based on the full pathway in VitisNet. The expression value of genes was normalized by DESeq2. Normalized expressions for control and ABA treatment groups are shown by red and blue lines respectively. Abbreviations: α-GAL, alpha-galactosidase; GolS, galactinol synthase; RafS, raffinose synthase; StaS: stachyose synthase. Refer to Fig. 2 and Fig. 3 for figure keys.
-
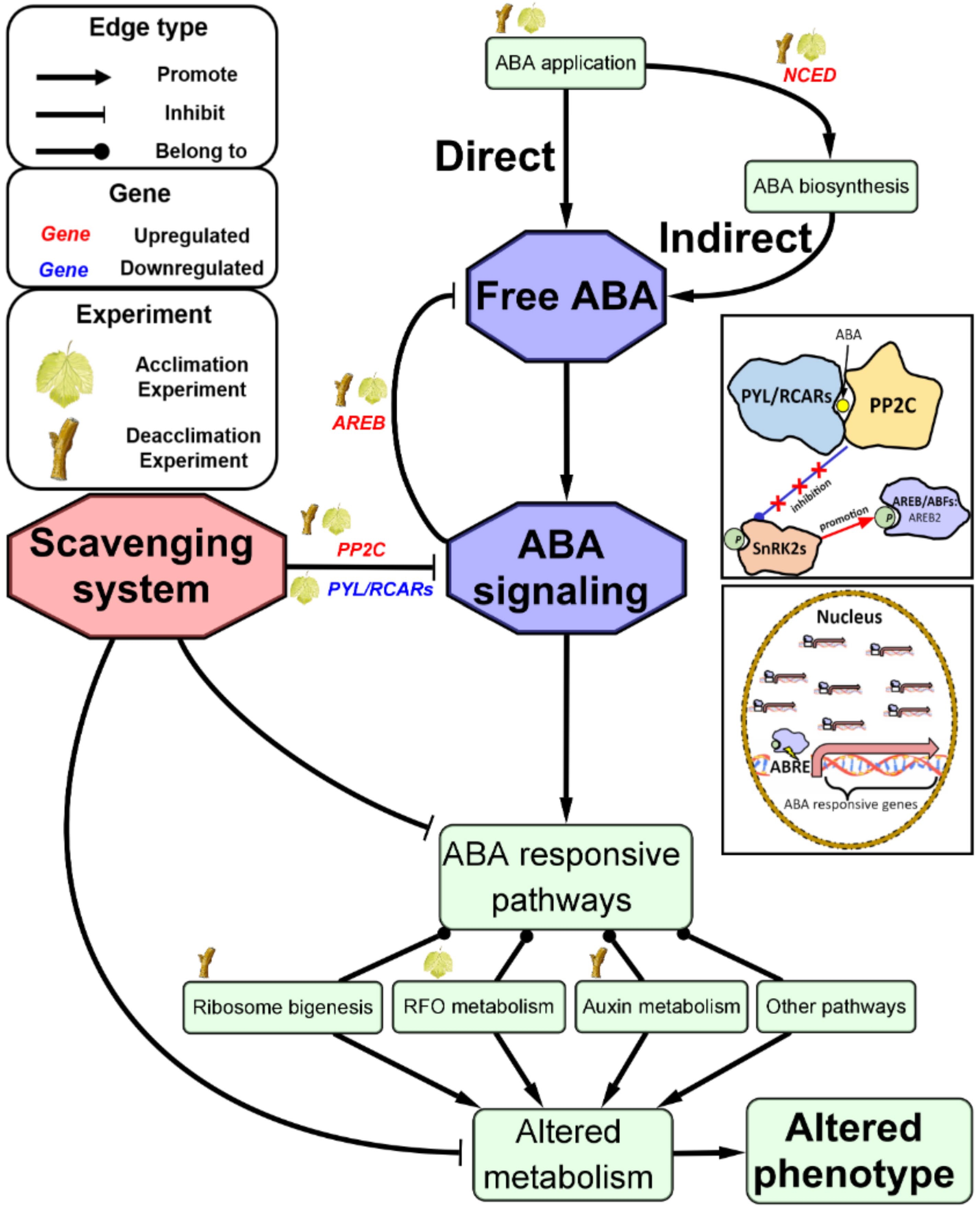
Figure 5. Schematic representation of a proposed model for exogenous ABA’s regulation on gene expression in grapevine cold acclimation and deacclimation.
Figures
(5)
Tables
(0)