-
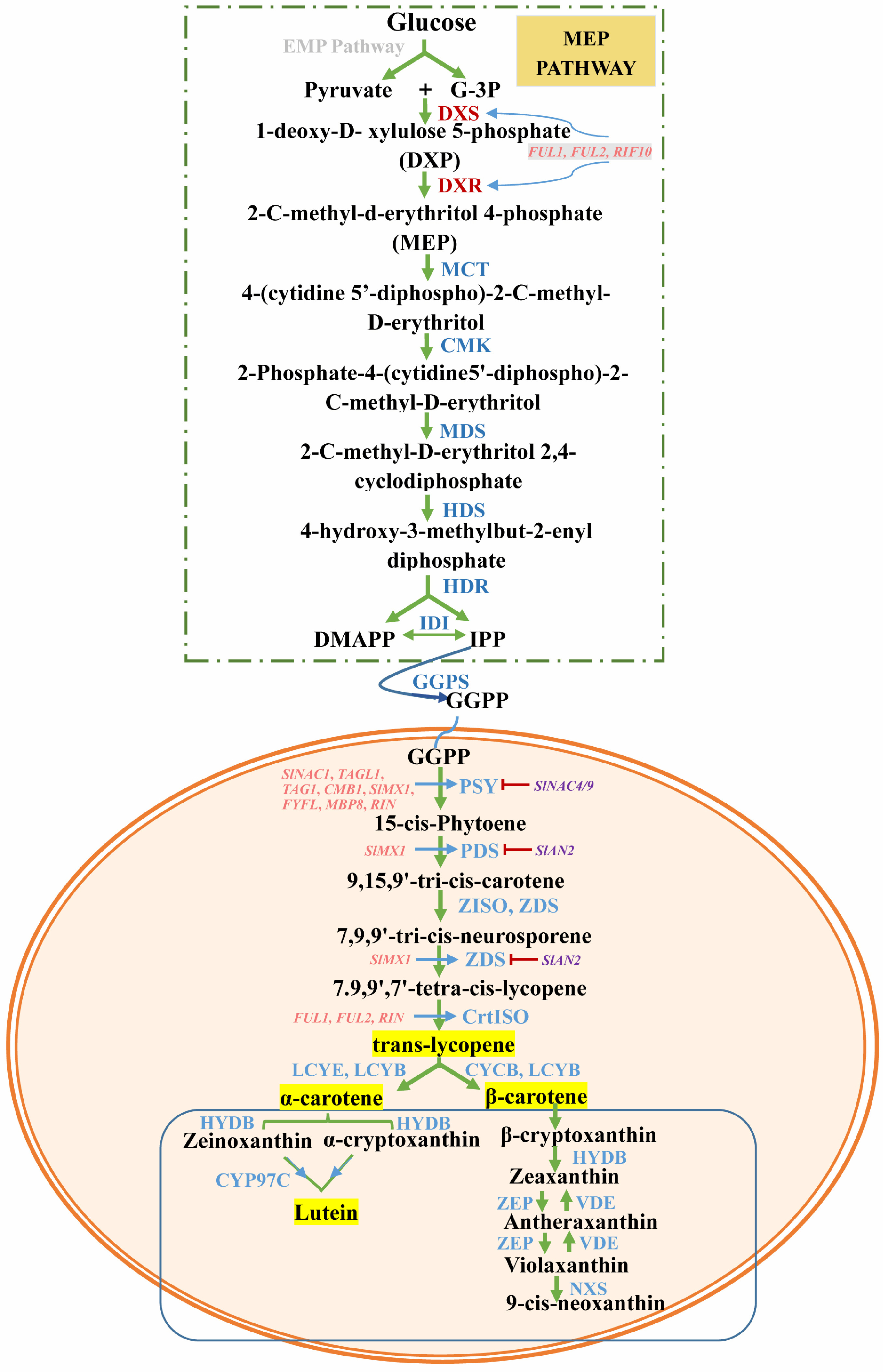
Figure 1.
The pathway of carotenoid biosynthesis and its transcriptional regulation. The pathway in the rectangle above represents the production of substrates for carotenoid synthesis through the MEP pathway. The pathway in the lower ellipsis represents the synthesis and decomposition of carotenoids in plastids. Enzymes are shown in blue. Blue arrows denote positive regulation, whereas red arrows denote negative regulation. DXS, 1-deoxy-D-xylulose-5-phosphate synthase; GGPP, geranylgeranyl pyrophosphate; DXR, 1-deoxy-d-xylulose 5-phosphate reductoisomerase; MCT, 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase; CMK, 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol kinase; MDS, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase; HDS, (E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase; HDR, (E)-4-hydroxy-3-methylbut-2-enyl diphosphate reductase; IDI, isopentenyl diphosphate isomerase; GPPS, geranyl diphosphate synthase; PSY, phytoene synthase; PDS, phytoene desaturase; ZDS, ζ-carotene desaturase gene; ZISO, ζ-carotene isomerase; CrtISO, carotene isomerase; LCY, lycopene beta-cyclase; HYDB, β-carotene hydroxylase; CYP97C, cytochrome P450-type monooxygenase 97C; VDE, violaxanthin de-epoxidase; ZEP, zeaxanthin epoxidase; NXS, neoxanthin synthase.
-
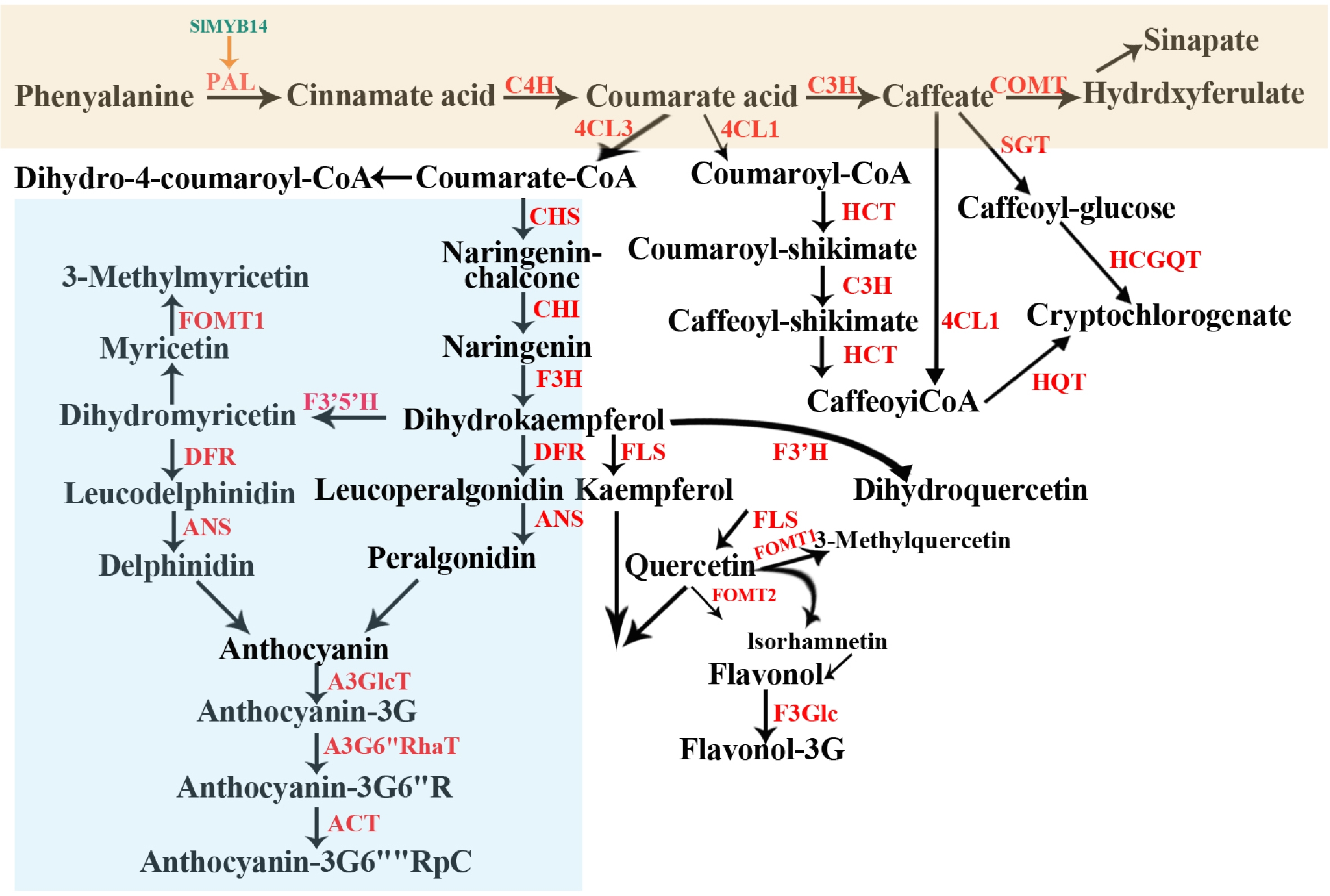
Figure 2.
Overview of the synthesis and metabolism framework of polyphenols in tomato. Enzymes are shown in red. Different colored rectangles represent the synthesis of different polyphenols. PAL, phenylalanine ammonia-lyase; C4H, cinnamate-4-hydroxylase; C3H, coumarate 3-hydroxylase; 4CL, 4-coumarate CoA ligase; COMT, catechol-Omethyl transferase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; DFR, dihydroflavonol reductase; ANS, anthocyanidin synthase; FOMT, flavonoid O-methyltransferase; F3'5'H, flavonoid-3,5-hydroxylase; A3GlcT, anthocyanin-3–O-glucosyltransferase; A3G6''RhaT, anthocyanin-3–O-glucoside-6′′–O-rhamnosyltransferase; ACT, Acylase; HCT, hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyltransferase; HCGQT, hydroxycinnamoyl glucose quinate hydroxycinnamoyl transferase; SGT, Solanidine: UDP-glucosyltransferase; HQT, hydroxycinnamoyl-Co A quinate hydroxycinamoyl transferase; F3'H, flavonoid 3-Hydroxylase; FLS, flavonol synthase.
-
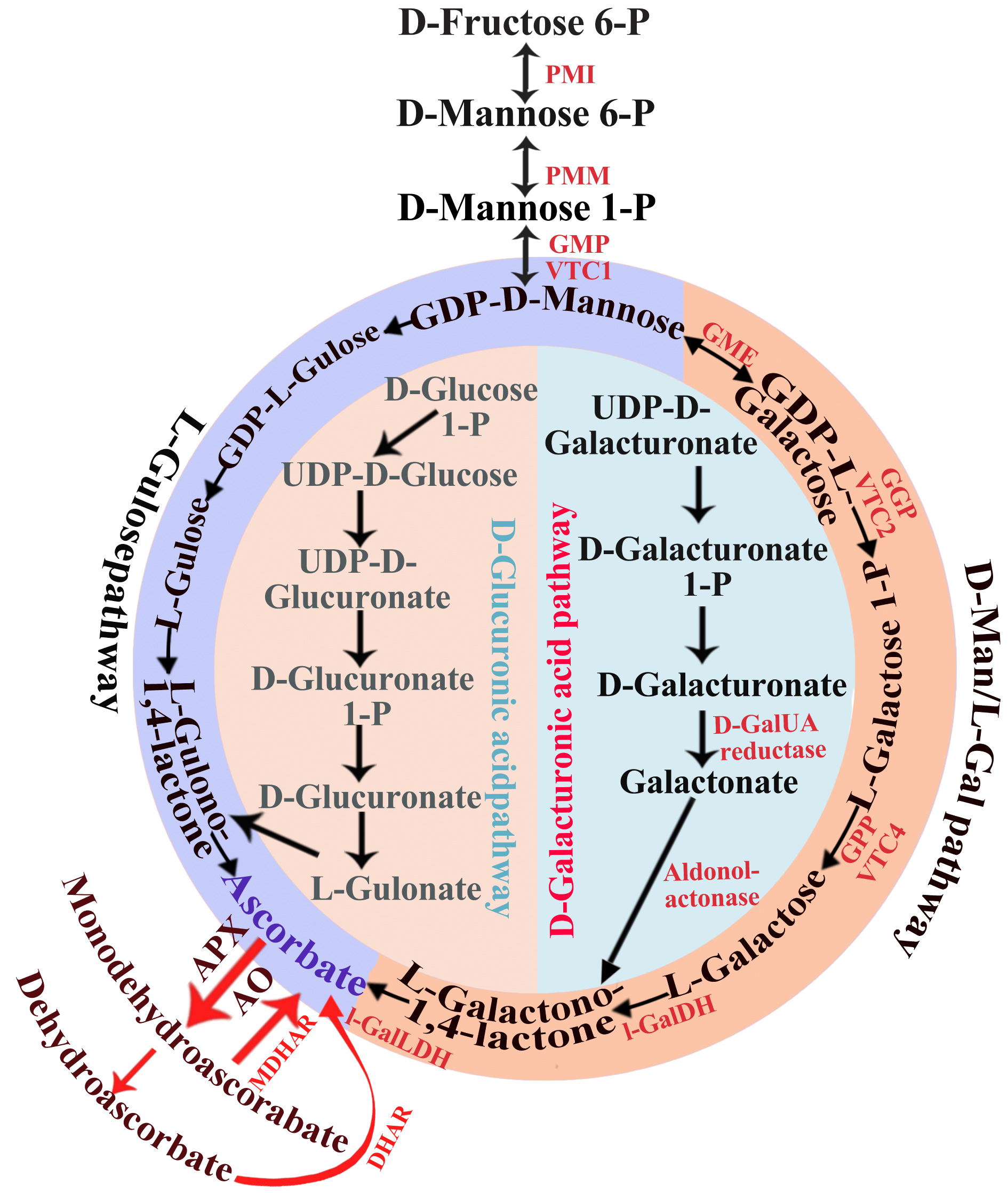
Figure 3.
Biosynthetic pathways of L-ascorbic acid in plants. PMI, mannose-6-phosphate isomerase; PMM, phosphomannomutase; GMP, GDP-mannose pyrophosphorylase (mannose-1-phosphate guanylyltransferase); GME, GDP-mannose-3′,5′-epimerase; GGP, GDP-L-galactose phosphorylase; L-GalDH, L-galactose dehydrogenase; L-GalLDH, L-galactono-1,4-lactone to produce ascorbate; AO, ascorbate oxidase; APX, ascorbate peroxidase; DHAR, dehydroascorbate reductase; MDHAR, monodehydroascorbate reductase; GPP, L-galactose-1-phosphate phosphatase.
-
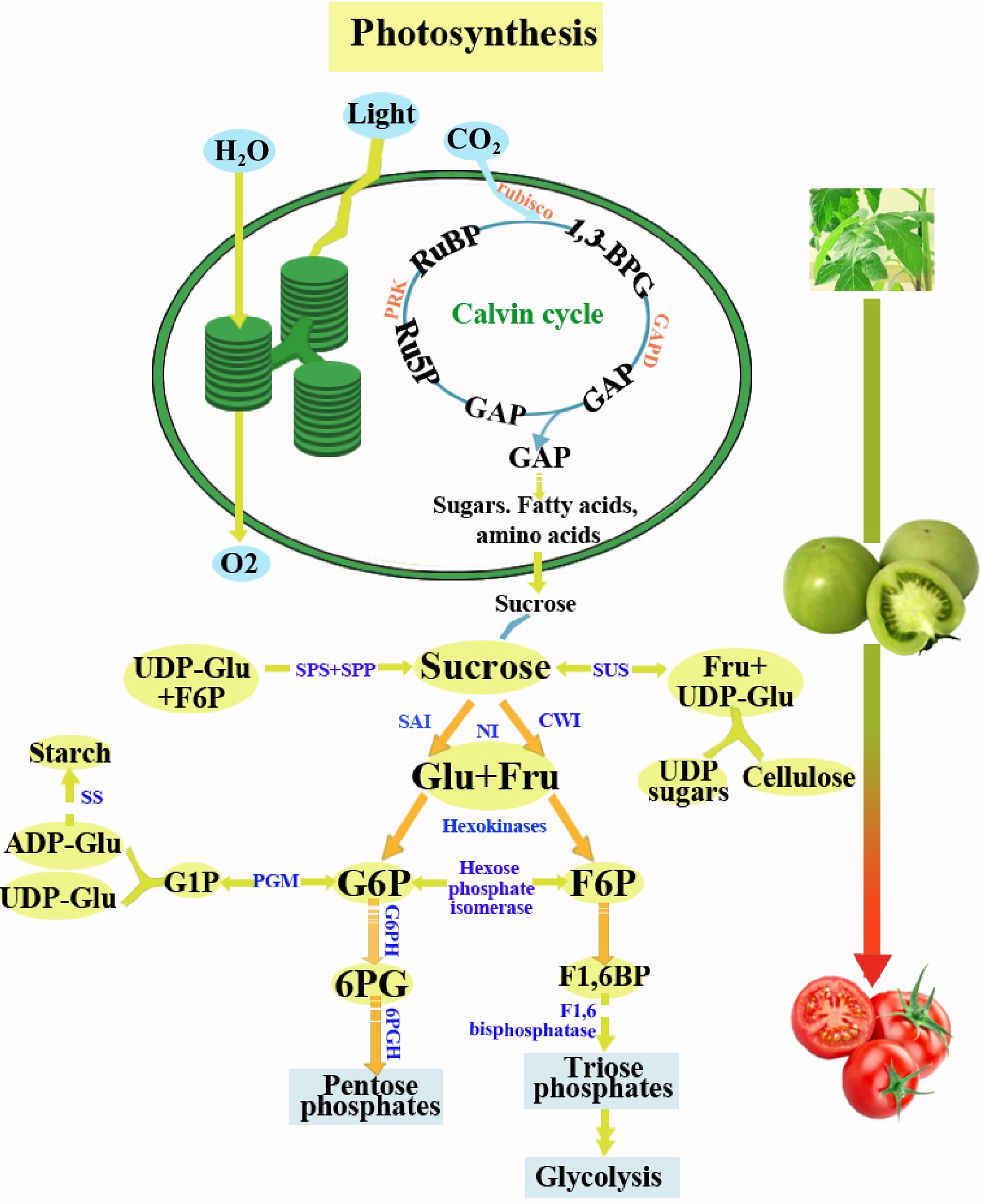
Figure 4.
Important intermediates in plant sugar metabolism. RuBP, ribulose-1,5-bisphosphate; 1,3-BPG, 1,3-bisphosphoglycerate; GAP, glyceraldehyde3-phosphate; Ru5P, Ribulose 5-phosphate; PRK, phosphoribulokinase; Glu, glucose; Fru, fructose; SAI, soluble acid invertase; NI, neutral invertase; CWI, cellwall invertase; G6P, glucose-6-phosphate; SUS, sucrose synthase; G1P, glucose-1-phosphate; F6P, fructose-6-phosphate; F1,6BP, fructose-1,6-bisphosphate; 6PG, 6-phosphogluconate; G6PH, glucose 6-phosphatedehydrogenase; 6PGH, 6-phosphogluconate dehydrogenase; PGM, Phosphoglucomutase; ADP-Glu, ADP-glucose; UDP-Glu, UDP-glucose; SS, Starch synthase; SPS, sucrose phosphate synthase; SPP, sucrose phosphate phosphatase;
-
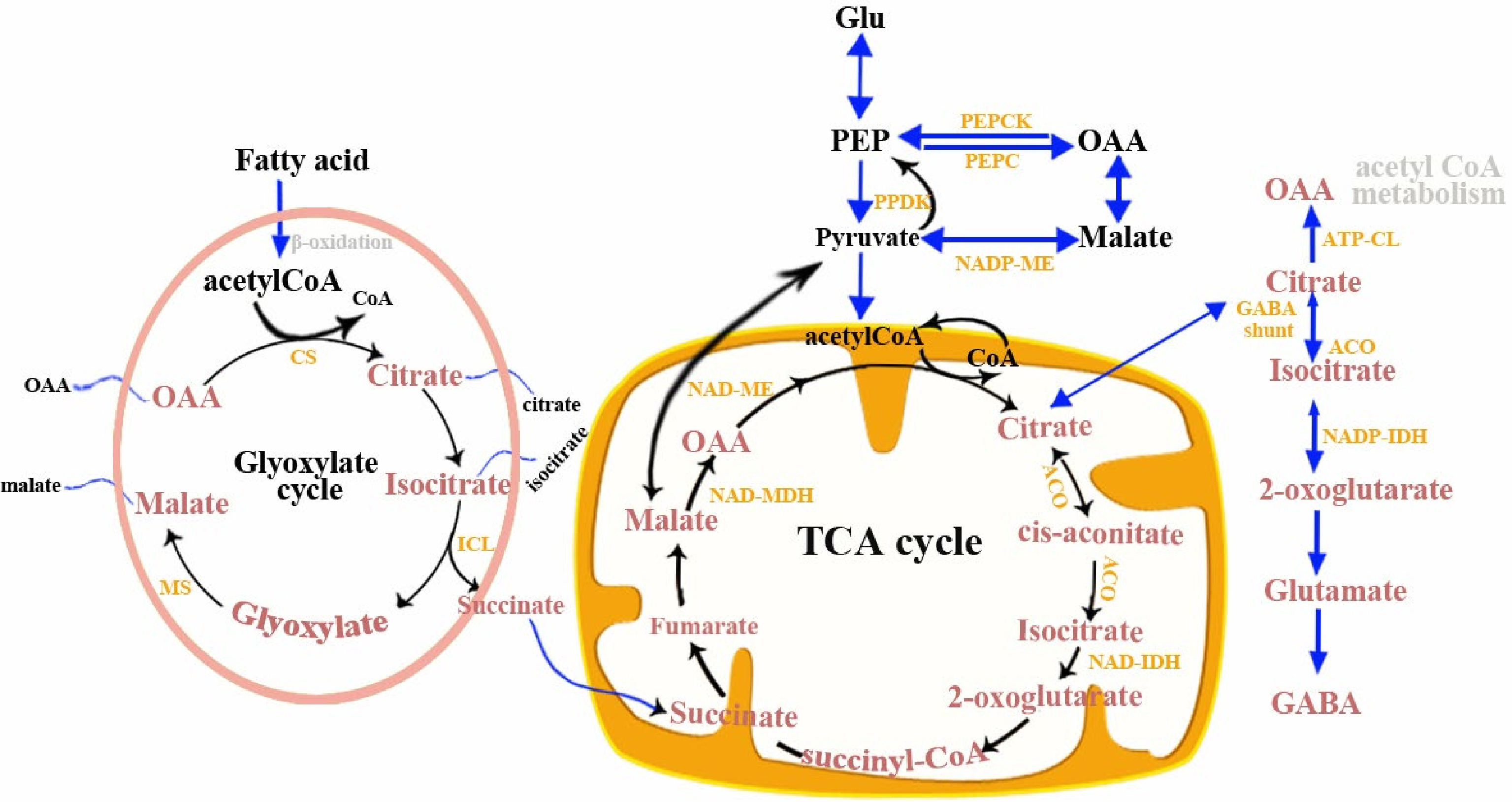
Figure 5.
Citric acid and malic acid metabolic pathways in fruit. TCA, tricarboxylic acid cycle; ACO, aconitase; ATP-CL, ATP-citrate lyase; CS, citrate synthase; ICL, isocitrate lyase; MS, malate synthase; NAD-MDH, NAD-malate; dehydrogenase; NAD-ME, NAD-malic enzyme; NAD-IDH, NAD-isocitrate dehydrogenase; NADP-ME, NADP-malic enzyme; OAA, oxaloacetate; NADP-IDH, NADP-isocitrate dehydrogenase; PDH, pyruvate dehydrogenase; PEPC, phosphoenolpyruvate carboxylase; PEPCK, phosphoenolpyruvate carboxykinase; PPDK, pyruvate orthophosphate dikinase.
-
Precursor Volatile compound Genes
(abbreviation)Full name Reference Fatty acid Hexanal TomloxC [72], [85] Trans-2-Heptenal 1-Penten-3-ol Hexanol Nonylaldehyde Heptaldehyde cis-3-Hexenol l-Penten-3-one Lsobutyl cyanide SlBCAT1 Tomato branched-chain amino acid aminotransferase [86] Trans-2-Pentenal SlscADH1 tomato short chain dehydrogenase-reductase [87] 3-Methylbutanol cis-3-hexenylacetate Hexylacetate Pentanol Isoleucine/Leucine 2/3-Methylbutanal SlscADH1 Tomato short chain dehydrogenase-reductase [87] Trans-2-Hexenal LeHPL Hydroperoxidelyase [88] Phenylalanine Methyl salicylate SlSAMT tomato salicylic acid methyl transferase [75] Phenylacetaldehyde LeAADC Aromatic amino acid decarboxylase [54] 2-Phenylethanol l-nitro-2-Phenylethane Benzylcyanide Benzaldehyde Methionine Eugenol Leucine 2-Isobutylthiazole lycopene β-Cyclocitral LeCCD4 Carotenoid cleavage dioxygenases [31] β-carotene β-ionone Carotenoid Pseudoionone LeCCD1B Carotenoid cleavage dioxygenases [31] ζ-caroteno Geranylacetone Carotenoid β-Damasccnonc Lycopene 6-Methyl-5-hepten-2-one Carotenoid Epoxy-B-ionone Geranial SlADH2 Tomato alcohol dehydrogenases [89] Neral Salicylaldehyde Trans-3-hexen-1-ol MiADH1 Mango alcohol dehydrogenase [90] Monoterpenoids Linalool SlGPPS, LeMTS1 Geranyl diphosphate synthases, linalool synthase [91] Table 1.
List of selected volatile compounds related to the flavor of tomatoes and the genes that regulate their accumulation.
-
Categories Genes
(abbreviation)Full name Function Reference Soluble sugar SlCIF1/2 Tomato cell wall invertase inhibitor gene It can inhibit the activity of CWIN at the post-translational level [60] INH1 Invertase inhibitor gene It can inhibit the activity of CWIN at the post-translational level SlHSP17.7 Tomato heat shock proteins encoding gene It can increase the accumulation of sugar [48] SlVPE1/2/3/4/5 Tomato vacuolar processing enzyme encoding gene It can promote acid invertases synthesis and improve hexose level [76] SlPEPCK Tomato phosphoenolpyruvate carboxykinase encoding gene It can regulate gluconeogenesis and enchance high sugar/acid ratio [77] LeLIN Fruit apoplastic invertase encoding gene Increase the activity of invertases [78] SlARF4 Abaxial encoding genes Inhibite the activity of AGPase and decreasing starch levels [79] TFT1, TFT10 14-3-3 protein encoding gene Down-regulated sucrose phosphate synthase activity [80] SlHXK1 Hexokinase encoding gene It can inhibit the accumulate of starch in tomato leaves [81] LeHT1/2/3 Tomato hexose transporter encoding gene Encode the hexose transporters and concentrate hexoses in storage parenchyma cells [82] SlSUT1 Tomato sucrose transporter encoding gene Phloem loader of sucrose in Solanaceae / RIN Ripening inhibitor Regulate sugar metabolism [61] SlMBP22 MADS-box subfamily, Bsister (Bs) genes It positively modulates the levels of starch and soluble sugar in tomato leaves [81] SlVIF Vacuolar invertase inhibitor gene It can inhibit the activity of VIN at the post-translational level [61] Organic acids SlPEPC2 Allosteric enzyme phosphoenolpyruvate carboxylase encoding gene Promote the decomposition of malic acid [83] TRXL1 Thioredoxin-like 1 encoding gene Activate NADP-MDH and increases malate [8] IDH3 Isocitrate dehydrogenases encoding gene Promote citric acid degradation [66] AST2 Aspartate aminotransferase encoding gene LeACS2 Ethylene biosynthesis gene Enhance the metabolization of malic and citric acid HMGCR1 3-hydroxy-3-methylglutaryl-CoA reductase encoding gene Inhibit the accumulation of critic acid MDH Malate dehydrogenase encoding gene Increase the content of malic acid PDC3 phosducin-like 3 encoding gene Decrease the content of malic acid SlAREB1 ABA-response element binding factor Mediate ABA signal and increase citric acid and malic acid level [84] AP2/ERF APETALA2/Ethylene Response Factor Regulate the organic acid levels [66] SlWRKYs Ethylene-responsive genes SNAC Stress-responsive NAC transcription factors Table 2.
Overview of the genes that affect the accumulation of soluble sugar and organic acids in tomatoes.
Figures
(5)
Tables
(2)