-
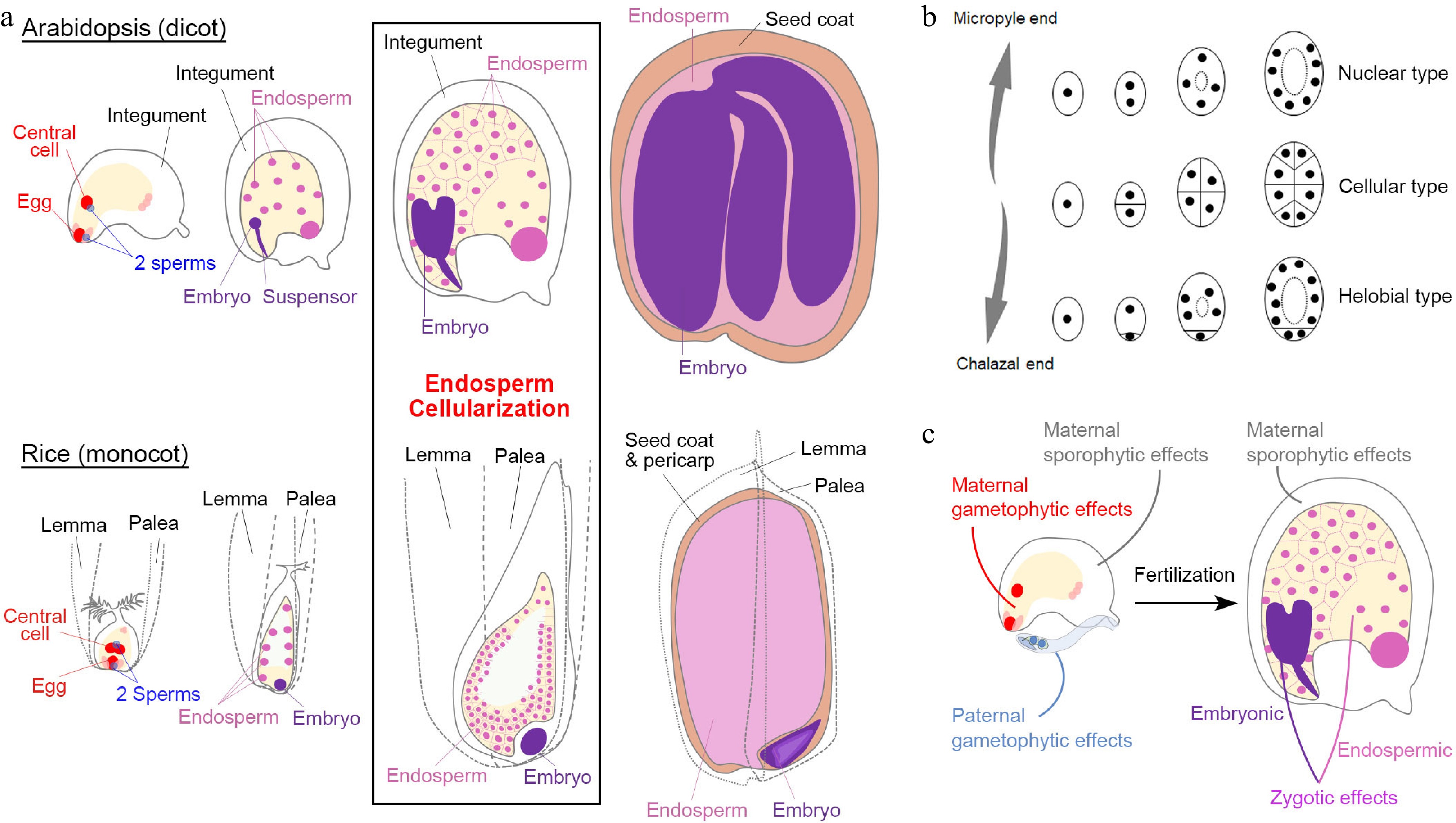
Figure 1.
Seed development in angiosperms. (a) Double fertilization (leftmost panels) initiates embryo and endosperm formation (right panels) across successive stages of seed development in Arabidopsis (dicot model) and rice (monocot model). (b) Different types of angiosperm endosperms. Dots denote endosperm nuclei, while ellipses denote the embryo sac before fertilization or the endosperm after fertilization. (c) Various maternal and paternal effects on the regulation of seed development. (a) & (c) Maternal and paternal components are indicated in red and blue, respectively. The seed coat is indicated in light brown. For the zygotic tissues, the endosperm and embryo are indicated in pink and purple, respectively.
-
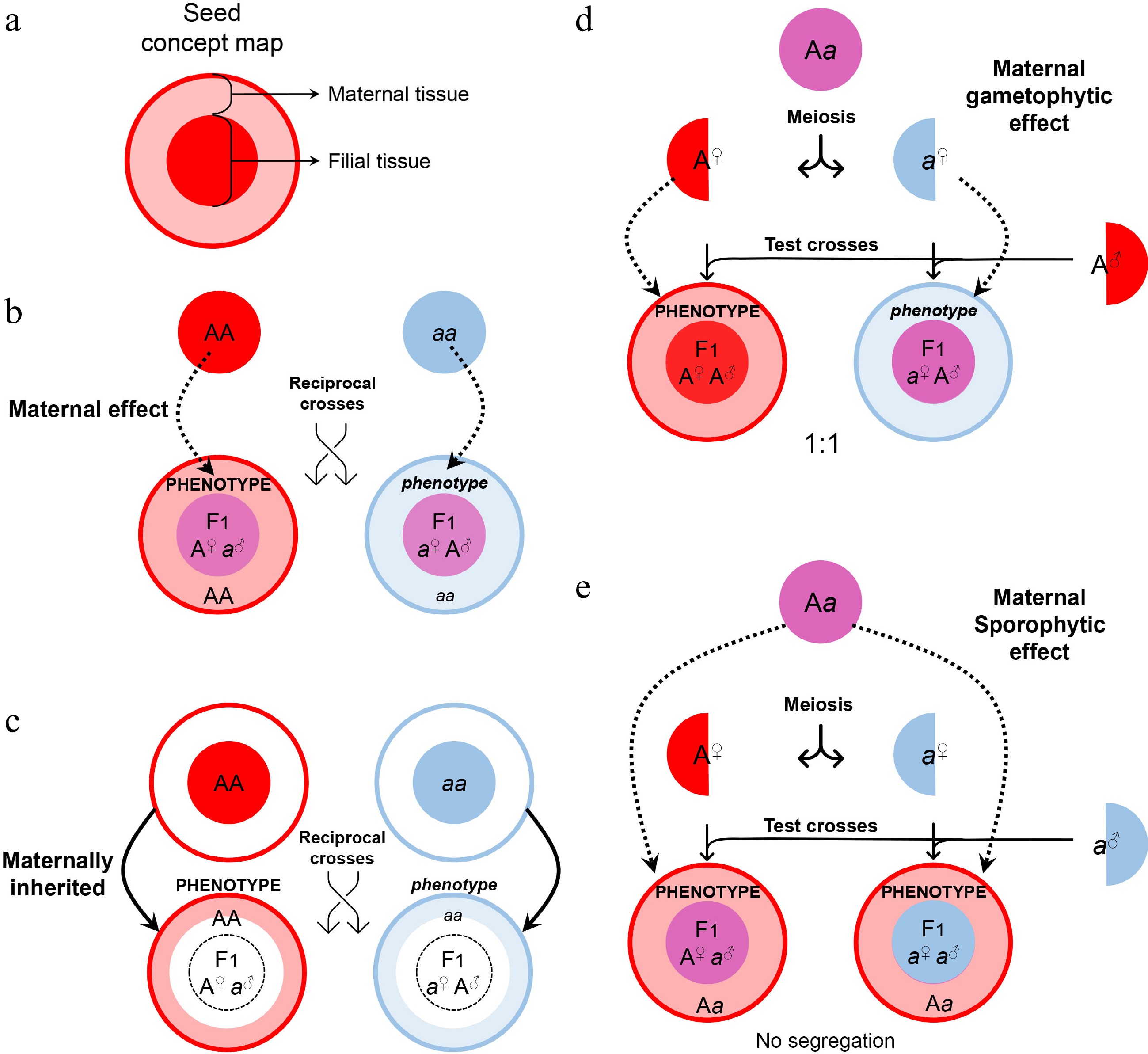
Figure 2.
Maternal control of seed development. (a) Symbols of maternal and filial tissues appearing in this figure. (b) Scheme of typical maternal effects. The phenotype of developing or mature seeds is determined by the maternal genotype. (c) Scheme of on-site effects of the maternal tissue. The phenotype is restricted to the tissue inherited from the mother, and thus determined by the maternal genotype. (d) Characterization of gametophytic maternal effects by test crosses. As the phenotype of developing or mature seeds is determined by the genotype of the female gametophyte, phenotypic segregation is observable in F1 progenies of test crosses. (e) Characterization of sporophytic maternal effects by test crosses. As the phenotype of developing or mature seeds is determined by the genotype of the female sporophyte, phenotypic segregation is unobservable in F1 progenies of test crosses.
-
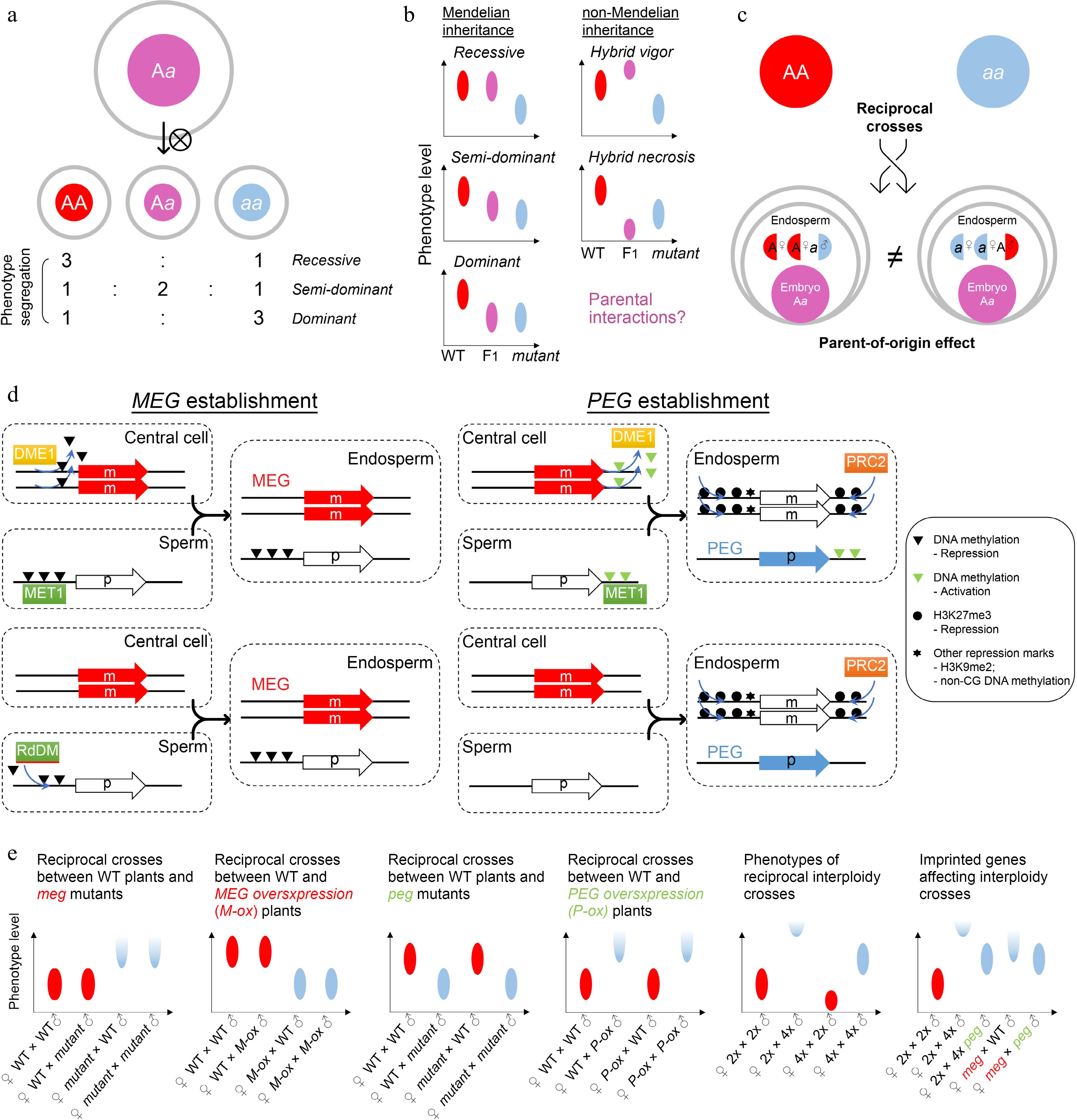
Figure 3.
Zygotic control of seed development. (a) A typical zygotic effect causes phenotype segregation among F1 siblings. (b) Scheme of Mendelian and non-Mendelian inheritance patterns of F1 progenies with zygotic effects. Left panels: possible phenotypes of F1 progenies with a recessive, semi-dominant, or dominant mutation. Right panels: possible phenotypes of F1 progenies with a non-mendelian mutation. Such patterns of non-mendelian inheritance are likely related to parental interactions. Ellipse indicates the quantified range of a phenotype. (c) Scheme of endospermic factors regulating F1 phenotypes in a parental-dependent manner. (d) Typical mechanisms underlying gene imprinting. MEG, maternally imprinted gene; PEG, paternally imprinted gene. (e) Phenotypic assumptions based on unbalanced parental dosage. The first four panels from the left show phenotypic patterns of reciprocal crosses between wild-type plants and plants with loss of function or overexpression of imprinted genes. The fifth panel shows phenotypic patterns of interploidy crosses in Arabidopsis, while the sixth panel shows a smilar phenotype between the paternal-excess cross (2nd column) and the cross with loss of MEG (meg) (4th column). Such phenotypes are suppressed by loss of PEG (peg) (3rd and 5th columns). Ellipse indicates the quantified range of a phenotype, while half ellipse indicates a possible abortive phenotype.
-
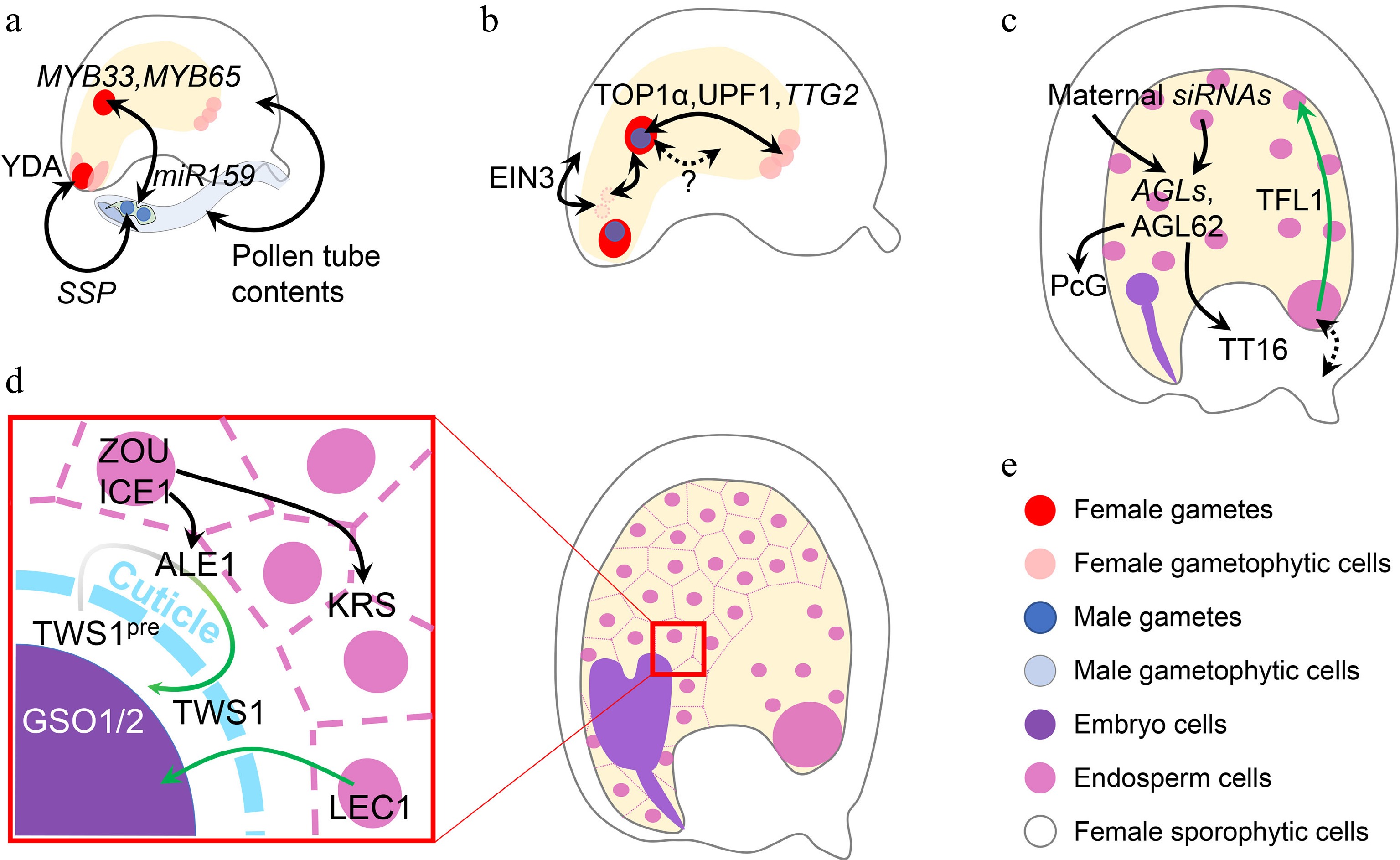
Figure 4.
Inter-tissue communication in seed development. (a) Inter-tissue communication at the beginning of seed development. Paternal SSP mRNA from the pollen affects the zygotic YDA pathway to determine zygote division. Paternal miR159 from the pollen quenches maternal MYB33/65 to initiate nascent endosperm division. Other contents in the pollen tube can also trigger ovule growth, which mimicks fertilization. (b) Inter-tissue communication in early seed development. Synergid nuclei affect the maternal-paternal genome ratio in the nascent endosperm, which is oppositely regulated by sporophytic and gametophytic EIN3. The communication between maternal antipodal cells and paternal cues in the nascent endosperm relies on the relative dosage of maternal and paternal TTG2, which is transcriptionally regulated by TOP1α and UPF1. Nascent endosperm-female gametophyte communication is also suggested, although the mechanisms are yet unknown. (c) Inter-tissue communication regulating endosperm and integument development. Chalazal-transcribed TFL1 functions in the peripheral endosperm to regulate endosperm cellularization. This module also infers a potential maternal-filial communication at the chalazal part. Endosperm regulators, AGLs, are regulated by maternal siRNAs from both the endosperm and maternal tissues. AGL62 in turn regulates maternal nucellus degradation via the maternal TT16 and integument growth via the maternal PcG complex. (d) Inter-tissue communication between endosperm and embryo. When the cuticle barrier between endosperm and embryo is not established, endosperm-expressed LEC1 relocates into the embryo to exert its function. The integrity of such a barrier is monitored by two-way communication, in which the precursor of the embryo-expressed TWS1 peptide (TWS1pre) is processed in the endosperm by ALE1 and the mature peptide signal moves back into the embryo to activate the GSO1/2-pathway. (e) Color legend shows different elements in this figure. Single- and double-headed arrows indicate one-way and reciprocal regulations, respectively. Dashed arrow indicates a putative regulation. Green single-headed arrow indicates protein movement, while green gradient single-headed arrow indicates protein movement along with the maturation process.
-
Gene name Abbreviation Gene ID Function note Reference ABERRANT TESTA SHAPE ATS AT5G42630 KANADI family transcription factor [30,31] ABNORMAL LEAF-SHAPE 1 ALE1 AT1G62340 Subtilisin-like serine protease [126−129] ABSCISIC ACID INSENSITIVE 3 ABI3 AT3G24650 B3 domain transcription factor [48−50] ADMETOS ADM AT4G11940 J-domain chaperone [99] ADRENODOXIN 1 ADX1 AT4G05450 Adrenodoxin [47] ADRENODOXIN 2 ADX2 AT4G21090 Adrenodoxin [47] ADRENODOXIN REDUCTASE ADXR AT4G32360 Adrenodoxin reductase [47] AGAMOUS-LIKE 40 AGL40 AT4G36590 MADS-box family transcription factor [32] AGAMOUS-LIKE 62 AGL62 AT5G60440 MADS-box family transcription factor [122] AGAMOUS-LIKE 91 AGL91 AT3G66656 MADS-box family transcription factor [32] AINTEGUMENTA ANT AT4G37750 AP2 family transcription factor [22, 23] APETALA2 AP2 AT4G36920 AP2 family transcription factor [29] BABY BOOM BBM AT5G17430 AP2 family transcription factor [51−53] CYTOCHROME P450 FAMILY 78 A7 CYP78A7 AT5G09970 Cytochrome p450 family [36,37] CYTOCHROME P450 FAMILY 78 A9 CYP78A9 AT3G61880 Cytochrome p450 family [36,37] DA1 DA1 AT1G19270 Ubiquitin-activated peptidase [26,27] DA2 DA2 AT1G78420 RING-type E3 ubiquitin ligase [26] DEMETER DME AT5G04560 DNA glycosylase [65,66,68,
69,78,80]DOSAGEEFFECT DEFECTIVE 1 DED1 Zm00001eb050770 MYB family transcription factor [93] ENDOSPERM BREAKDOWN1 ENB1 Zm00001eb061800 Cellulose synthase 5 [62] ENHANCER OF da1-1 3 EOD3 (CYP78A6) AT2G46660 Cytochrome p450 family [36] ETHYLENE INSENSITIVE 3 EIN3 AT3G20770 Transcription regulator [114] FERTILIZATION INDEPENDENT SEED 2 FIS2 AT2G35670 PRC2 component [72,89] FLOWERING WAGENINGEN FWA AT4G25530 Homeodomain-containing transcription factor [73] FUSCA3 FUS3 AT3G26790 B3 domains transcription factor [48−50] GASSHO1 GSO1 AT4G20140 Leucine rich repeat (LRR) receptor-like kinase [126,127] GASSHO2 GSO2 AT5G44700 Leucine rich repeat (LRR) receptor-like kinase [126,127] GIANT EMBRYO GE (OsCYP78A13) LOC_Os07g41240 Cytochrome p450 family [45,46] GLABRA2 GL2 AT1G79840 Homeodomain-containing transcription factor [17] GRAIN WEIGHT 2 GW2 LOC_Os02g14720 RING-type E3 ubiquitin ligase [41] HAIKU1 IKU1 AT1G55600 Plant-specific VQ motif-containing protein [5,6,8] HAIKU2 IKU2 AT3G19700 Leucine rich repeat (LRR) kinase [5−7] HOMEDOMAIN GLABROUS 3 HDG3 AT2G32370 Homeodomain-containing transcription factor [92] INDUCER OF CBF EXPRESSIONICE 1 ICE1 AT3G26744 bHLH family transcription factor [128, 129] INNER NO OUTER INO AT1G23420 YABBY family transcription factor [24] KERBEROS KRS AT1G50650 STIG1 family of peptide [130] KLUH KLU (CYP78A5) AT1G13710 Cytochrome p450 family [35] LEAFY COTYLEDON 1 LEC1 AT1G21970 Nuclear factor Y transcription factor [48−50] LEAFY COTYLEDON 2 LEC2 AT1G28300 B3 domains transcription factor [48−50] MATERNAL DEREPRESSION OF r1 MDR1 (DNG101) Zm00001eb202980 DNA glycosylase [81] MATERNAL EFFECT EMBRYO ARREST45 MEE45 AT4G00260 B3 domains transcription factor [38] MATERNALLY EXPRESSED PAB C-TERMINAL MPC AT3G19350 C-terminal domain of poly(A) binding protein [71] MEDEA MEA AT1G02580 PRC2 component [69,72,74,75,
83,84,89]METHYLTRANSFERASE 1 MET1 AT5G49160 Methyltransferase 1 [70, 83,84,
86,106]MINISEED3 MINI3 AT1G55600 WRKY family transcription factor, WRKY10 [6,7] MIR159a MIR159a AT1G73687 MicroRNA [112] MIR159b MIR159b AT1G18075 MicroRNA [112] MIR159c MIR159c AT2G46255 MicroRNA [112] MYB33 MYB33 AT5G06100 MYB family transcription factor [112] MYB65 MYB65 AT3G11440 MYB family transcription factor [112] PHERES 1 PHE1(AGL37) AT1G65330 MADS-box family transcription factor [83−85] PHOSPHATE 1 PHO1 AT3G23430 Phosphate transporter [19] PICKLE RELATED 2 PKR2 AT4G31900 Chromatin remodeling factor [104] OsBBM1 OsBBM1 LOC_Os11g19060 AP2 family transcription factor [110,111] SHAGGY-LIKE KINASE 11 SK11 AT5G26751 GSK3 family/SHAGGY-like protein kinase [16, 18] SHAGGY-LIKE KINASE 12 SK12 AT3G05840 GSK3 family/SHAGGY-like protein kinase [16,18] SHORT HYPOCOTYL UNDER BLUE1 SHB1 AT4G25350 homologous with SYG1 protein family members, transcription regulator [9] SHORT SUSPENSOR SSP AT2G17090 Receptor-like cytoplasmic protein kinase [108,109] SmD1b SmD1b AT4G02840 Smith protein [18] TERMINAL FLOWER1 TFL1 AT5G03840 Phosphatidylethanolamine binding protein (PEBP) family member [121] TOPOISOMERASE Iα TOP1α AT5G55300 DNA topoisomerase [117] TRANSPARENT TESTA 16 TT16 (AGL32) AT5G23260 MADS-box family transcription factor [123] TRANSPARENT TESTA 2 TT2 AT5G35550 MYB family transcription factor [13] TRANSPARENT TESTA 8 TT8 AT4G09820 bHLH family transcription factor [14] TRANSPARENT TESTA GLABRA 1 TTG1 AT5G24520 WD40-motif containing transcription regulator [15, 16] TRANSPARENT TESTA GLABRA 2 TTG2 AT2G37260 WRKY family transcription factor, WRKY44 [28,117] TWISTED SEED 1 TWS1 AT5G01075 Signaling peptide precursor [126, 127] UBIQUITIN-SPECIFIC PROTEASE 12 UBP12 AT5G06600 Deubiquitination enzyme [27] UBIQUITIN-SPECIFIC PROTEASE 13 UBP13 AT3G11910 Deubiquitination enzyme [27] UP-FRAMESHIFT SUPPRESSOR 1 UPF1 AT5G47010 RNA helicase [117] YODA YDA AT1G63700 Member of MEKK subfamily, involved in MAPK cascade [108,109] ZHOUPI ZOU AT1G49770 bHLH family transcription factor [128, 129] ZmGW2-CHR4 ZmGW2-CHR4 Zm00001eb204560 RING-type E3 ubiquitin ligase [43] ZmGW2-CHR5 ZmGW2-CHR5 Zm00001eb238650 RING-type E3 ubiquitin ligase [43] ZmSWEET4c ZmSWEET4c Zm00001eb236820 Sugar transporter [20] Table 1.
Information on the genes discussed in this review.
Figures
(4)
Tables
(1)