-
Gymnosperms and angiosperms are called seed plants because they reproduce by seeds. In particular, the angiosperms, also known as flowering plants or higher plants, are the most diverse and widespread group of land plants on earth[1]. As indicated by their names, gymnosperm seeds are exposed without the protection layer, while angiosperm seeds are embedded in the maternal fruit. The seed origin of angiosperms is also different from that of gymnosperms. Angiosperm seeds result from a unique double fertilization process, in which one sperm nucleus (haploid; 1n) fertilizes the egg cell (haploid; 1n) and another sperm nucleus fertilizes the central cell (either 1n + 1n or 2n). The sperm-egg fusion produces the embryo (diploid; 2n), while the sperm-central cell fusion develops into the endosperm (triploid; 3n), which is an angiosperm-specific and terminally differentiated tissue that provides nutrition to the embryo or young seedling (Fig. 1a).
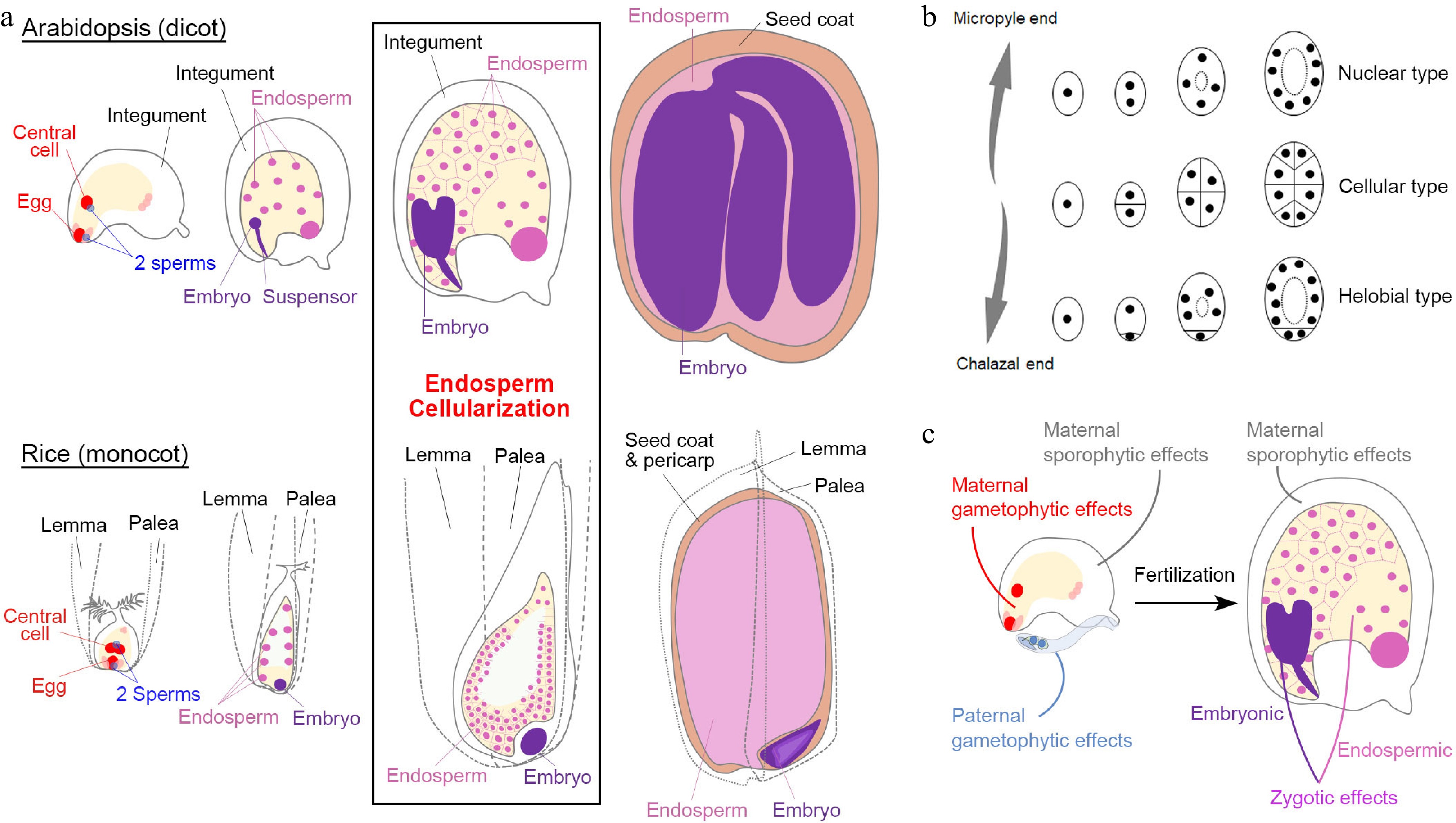
Figure 1.
Seed development in angiosperms. (a) Double fertilization (leftmost panels) initiates embryo and endosperm formation (right panels) across successive stages of seed development in Arabidopsis (dicot model) and rice (monocot model). (b) Different types of angiosperm endosperms. Dots denote endosperm nuclei, while ellipses denote the embryo sac before fertilization or the endosperm after fertilization. (c) Various maternal and paternal effects on the regulation of seed development. (a) & (c) Maternal and paternal components are indicated in red and blue, respectively. The seed coat is indicated in light brown. For the zygotic tissues, the endosperm and embryo are indicated in pink and purple, respectively.
Except for basal angiosperm species, the majority of angiosperms can be roughly divided into two groups, monocots and dicots, which exhibit various features of the seed structure. Some dicot seeds, including those of the model plant Arabidopsis (Arabidopsis thaliana), bear degraded endosperms as the nutrients are mainly stored in mature embryos (Fig. 1a). In the monocot Poaceae family, including popular crops such as rice (Oryza sativa), maize (Zea mays), barley (Hordeum vulgare), and wheat (Triticum aestivum), the commonly termed 'seed' is the caryopsis (a kind of fruit), in which the seed coat is fused to the pericarp (the fruit coat). The mature seeds of these crops have well-developed endosperms that store nutrients (Fig. 1a). Although seed development in these monocot crops coincides with fruit development in nature, their basic stages are comparable with those in dicots[2] (Fig. 1a).
The embryogenesis process is geometrically different in dicots and monocots[2]. For example, the first zygotic division in Arabidopsis is asymmetric, resulting in a small apical cell and a large basal cell. The cell lineage from the basal cell generates the suspensor and part of the embryonic root apical meristem, while the other embryonic tissues generate from the apical cell. In contrast, the rice zygote undergoes random divisions to generate a cluster of cells before differentiation (Fig. 1a), indicating that establishment of embryonic patterning is much later in monocots than in dicots. Moreover, embryonic differentiation in Arabidopsis is along the apical-basal axis with bilateral symmetry, whereas embryonic differentiation in rice exhibits an evident dorsal-ventral axis with both shoot and root meristem cells occurring at the ventral side.
As the featured structure of angiosperm seeds, endosperms are classified into three types: nuclear type, cellular type, and helobial type[3]. The nuclear-type endosperm is the most common type in which the primary endosperm undergoes karyokinesis repeatedly without cell wall formation to produce free nuclei at earlier stages. The cell wall only appears during endosperm cellularization to separate individual nuclei (Fig. 1a & b). In contrast, the cellular-type endosperm proliferates via complete cytokinesis with cell wall formation from the very beginning (Fig. 1b). The helobial-type endosperm is an intermediate type in which the chalazal endosperm undergoes complete cytokinesis once or twice, while the micropyle endosperm undergoes karyokinesis (Fig. 1b).
Both Arabidopsis and rice develop the nuclear-type endosperm (Fig. 1a & b). In Arabidopsis, endosperm cellularization occurs during the embryo status at the heart stage to the early torpedo stage, except that its chalazal endosperm never undergoes cellularization[2]. After endosperm cellularization, endosperm cells undergo endoreplication in Arabidopsis, whereas in monocots, numerous additional rounds of mitoses occur between endosperm cellularization and endoreduplication[4]. Endosperm cellularization is crucial for seed development[5−9]. Generally, the over-proliferated endosperm is associated with delayed or failed cellularization, resulting in larger or aborted seeds, respectively. In contrast, less-proliferated and accelerated endosperm cellularization leads to smaller seeds. At the end of seed development, the Arabidopsis endosperm is almost consumed by the embryo except for a one-cell layer adjacent to the seed coat, whereas the mature rice endosperm takes the major volume of a seed and differentiates into several functional regions[2]. Notably, although endosperm and embryo development are closely correlated, their individual development can proceed autonomously albeit defectively when the accompanying part is completely lost[10,11], indicating that the endosperm and embryo develop both independently and dependently.
In general, a mature angiosperm seed contains at least the diploid seed coat (parent generation; maternal sporophytic tissue), the diploid embryo (filial generation), and the triploid endosperm (filial generation) (Fig. 1a). Such heterogeneity implies that seed development is regulated by interwoven signaling networks. Before fertilization, maternal and paternal gametophytic effects influence the formation of gametophytic embryo sac and pollen prior to seed development. After fertilization, factors pertaining to filial tissues (zygotic tissues) could play a more specific role in embryonic or endospermic development. Because the 'paternal sporophyte' is not involved in seed development, a paternal effect is equivalent to the paternal gametophytic effect and zygotic paternal effect (sometimes known as xenia effects[12]). Notably, since seed development depends on the maternal support in the course of the whole seed developmental process, the maternal sporophytic effects play pivotal roles both before and after fertilization (Fig. 1c). In the following sections, we discuss parental effects, including paternal effects, in the context of maternal, zygotic, and inter-tissue regulation. The genes discussed in these sections are summarized in Table 1.
Table 1. Information on the genes discussed in this review.
Gene name Abbreviation Gene ID Function note Reference ABERRANT TESTA SHAPE ATS AT5G42630 KANADI family transcription factor [30,31] ABNORMAL LEAF-SHAPE 1 ALE1 AT1G62340 Subtilisin-like serine protease [126−129] ABSCISIC ACID INSENSITIVE 3 ABI3 AT3G24650 B3 domain transcription factor [48−50] ADMETOS ADM AT4G11940 J-domain chaperone [99] ADRENODOXIN 1 ADX1 AT4G05450 Adrenodoxin [47] ADRENODOXIN 2 ADX2 AT4G21090 Adrenodoxin [47] ADRENODOXIN REDUCTASE ADXR AT4G32360 Adrenodoxin reductase [47] AGAMOUS-LIKE 40 AGL40 AT4G36590 MADS-box family transcription factor [32] AGAMOUS-LIKE 62 AGL62 AT5G60440 MADS-box family transcription factor [122] AGAMOUS-LIKE 91 AGL91 AT3G66656 MADS-box family transcription factor [32] AINTEGUMENTA ANT AT4G37750 AP2 family transcription factor [22, 23] APETALA2 AP2 AT4G36920 AP2 family transcription factor [29] BABY BOOM BBM AT5G17430 AP2 family transcription factor [51−53] CYTOCHROME P450 FAMILY 78 A7 CYP78A7 AT5G09970 Cytochrome p450 family [36,37] CYTOCHROME P450 FAMILY 78 A9 CYP78A9 AT3G61880 Cytochrome p450 family [36,37] DA1 DA1 AT1G19270 Ubiquitin-activated peptidase [26,27] DA2 DA2 AT1G78420 RING-type E3 ubiquitin ligase [26] DEMETER DME AT5G04560 DNA glycosylase [65,66,68,
69,78,80]DOSAGEEFFECT DEFECTIVE 1 DED1 Zm00001eb050770 MYB family transcription factor [93] ENDOSPERM BREAKDOWN1 ENB1 Zm00001eb061800 Cellulose synthase 5 [62] ENHANCER OF da1-1 3 EOD3 (CYP78A6) AT2G46660 Cytochrome p450 family [36] ETHYLENE INSENSITIVE 3 EIN3 AT3G20770 Transcription regulator [114] FERTILIZATION INDEPENDENT SEED 2 FIS2 AT2G35670 PRC2 component [72,89] FLOWERING WAGENINGEN FWA AT4G25530 Homeodomain-containing transcription factor [73] FUSCA3 FUS3 AT3G26790 B3 domains transcription factor [48−50] GASSHO1 GSO1 AT4G20140 Leucine rich repeat (LRR) receptor-like kinase [126,127] GASSHO2 GSO2 AT5G44700 Leucine rich repeat (LRR) receptor-like kinase [126,127] GIANT EMBRYO GE (OsCYP78A13) LOC_Os07g41240 Cytochrome p450 family [45,46] GLABRA2 GL2 AT1G79840 Homeodomain-containing transcription factor [17] GRAIN WEIGHT 2 GW2 LOC_Os02g14720 RING-type E3 ubiquitin ligase [41] HAIKU1 IKU1 AT1G55600 Plant-specific VQ motif-containing protein [5,6,8] HAIKU2 IKU2 AT3G19700 Leucine rich repeat (LRR) kinase [5−7] HOMEDOMAIN GLABROUS 3 HDG3 AT2G32370 Homeodomain-containing transcription factor [92] INDUCER OF CBF EXPRESSIONICE 1 ICE1 AT3G26744 bHLH family transcription factor [128, 129] INNER NO OUTER INO AT1G23420 YABBY family transcription factor [24] KERBEROS KRS AT1G50650 STIG1 family of peptide [130] KLUH KLU (CYP78A5) AT1G13710 Cytochrome p450 family [35] LEAFY COTYLEDON 1 LEC1 AT1G21970 Nuclear factor Y transcription factor [48−50] LEAFY COTYLEDON 2 LEC2 AT1G28300 B3 domains transcription factor [48−50] MATERNAL DEREPRESSION OF r1 MDR1 (DNG101) Zm00001eb202980 DNA glycosylase [81] MATERNAL EFFECT EMBRYO ARREST45 MEE45 AT4G00260 B3 domains transcription factor [38] MATERNALLY EXPRESSED PAB C-TERMINAL MPC AT3G19350 C-terminal domain of poly(A) binding protein [71] MEDEA MEA AT1G02580 PRC2 component [69,72,74,75,
83,84,89]METHYLTRANSFERASE 1 MET1 AT5G49160 Methyltransferase 1 [70, 83,84,
86,106]MINISEED3 MINI3 AT1G55600 WRKY family transcription factor, WRKY10 [6,7] MIR159a MIR159a AT1G73687 MicroRNA [112] MIR159b MIR159b AT1G18075 MicroRNA [112] MIR159c MIR159c AT2G46255 MicroRNA [112] MYB33 MYB33 AT5G06100 MYB family transcription factor [112] MYB65 MYB65 AT3G11440 MYB family transcription factor [112] PHERES 1 PHE1(AGL37) AT1G65330 MADS-box family transcription factor [83−85] PHOSPHATE 1 PHO1 AT3G23430 Phosphate transporter [19] PICKLE RELATED 2 PKR2 AT4G31900 Chromatin remodeling factor [104] OsBBM1 OsBBM1 LOC_Os11g19060 AP2 family transcription factor [110,111] SHAGGY-LIKE KINASE 11 SK11 AT5G26751 GSK3 family/SHAGGY-like protein kinase [16, 18] SHAGGY-LIKE KINASE 12 SK12 AT3G05840 GSK3 family/SHAGGY-like protein kinase [16,18] SHORT HYPOCOTYL UNDER BLUE1 SHB1 AT4G25350 homologous with SYG1 protein family members, transcription regulator [9] SHORT SUSPENSOR SSP AT2G17090 Receptor-like cytoplasmic protein kinase [108,109] SmD1b SmD1b AT4G02840 Smith protein [18] TERMINAL FLOWER1 TFL1 AT5G03840 Phosphatidylethanolamine binding protein (PEBP) family member [121] TOPOISOMERASE Iα TOP1α AT5G55300 DNA topoisomerase [117] TRANSPARENT TESTA 16 TT16 (AGL32) AT5G23260 MADS-box family transcription factor [123] TRANSPARENT TESTA 2 TT2 AT5G35550 MYB family transcription factor [13] TRANSPARENT TESTA 8 TT8 AT4G09820 bHLH family transcription factor [14] TRANSPARENT TESTA GLABRA 1 TTG1 AT5G24520 WD40-motif containing transcription regulator [15, 16] TRANSPARENT TESTA GLABRA 2 TTG2 AT2G37260 WRKY family transcription factor, WRKY44 [28,117] TWISTED SEED 1 TWS1 AT5G01075 Signaling peptide precursor [126, 127] UBIQUITIN-SPECIFIC PROTEASE 12 UBP12 AT5G06600 Deubiquitination enzyme [27] UBIQUITIN-SPECIFIC PROTEASE 13 UBP13 AT3G11910 Deubiquitination enzyme [27] UP-FRAMESHIFT SUPPRESSOR 1 UPF1 AT5G47010 RNA helicase [117] YODA YDA AT1G63700 Member of MEKK subfamily, involved in MAPK cascade [108,109] ZHOUPI ZOU AT1G49770 bHLH family transcription factor [128, 129] ZmGW2-CHR4 ZmGW2-CHR4 Zm00001eb204560 RING-type E3 ubiquitin ligase [43] ZmGW2-CHR5 ZmGW2-CHR5 Zm00001eb238650 RING-type E3 ubiquitin ligase [43] ZmSWEET4c ZmSWEET4c Zm00001eb236820 Sugar transporter [20] -
A maternal effect is generally defined as a phenomenon in which the offspring phenotype is determined by the genotype of its mother. However, the angiosperm seeds bear mixed features of two generations, which makes the maternal effects of angiosperms more complicated than those of animals.
Usually, maternal effects can be observed from reciprocal crosses, where the F1 progenies that have the same genetic background might show different phenotypes from the mother (Fig. 2a & b). For example, several TRANSPARENT TESTA (TT) genes in Arabidopsis, including TT2[13], TT8[14], and TRANSPARENT TESTA GLABRA1 (TTG1)[15,16], show maternal effects on the accumulation of seed oil in F1 progenies. The seeds from tt mutants pollinated with wild-type and its own pollen have similarly higher levels of seed oil than wild-type seeds, whereas the seeds from wild-type plants pollinated with tt pollen do not exhibit such a phenotype. Likewise, the regulators acting upstream and downstream of TTG1, including SmD1b, SHAGGY-LIKE KINASE 11/12 (SK11/12), and GLABRA2 (GL2), show maternal effects on regulating seed oil levels[16−18]. In addition, phosphate (Pi) exporters localized in the chalazal seed coat are crucial for Pi flux between the chalazal seed coat and the embryo, and such a remote control is evidenced by grafting assays[19]. Sugar transporters expressed in the maternal seed coat are responsible for transferring hexoses across the basal endosperm transfer layer to the starch-storing endosperm in rice and maize[20]. These findings suggest that maternal effects play a central regulatory role in relocating resources into seeds.
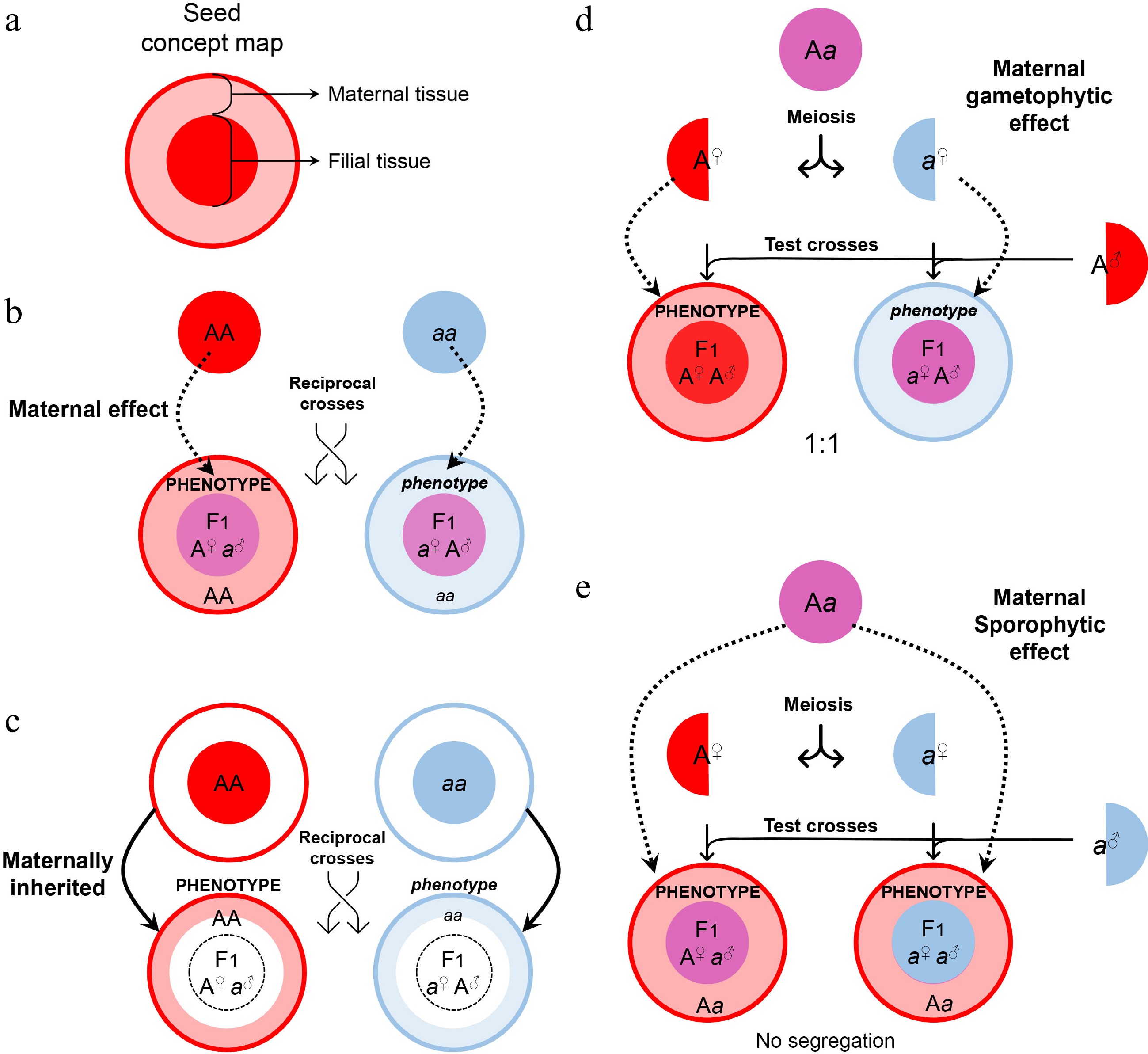
Figure 2.
Maternal control of seed development. (a) Symbols of maternal and filial tissues appearing in this figure. (b) Scheme of typical maternal effects. The phenotype of developing or mature seeds is determined by the maternal genotype. (c) Scheme of on-site effects of the maternal tissue. The phenotype is restricted to the tissue inherited from the mother, and thus determined by the maternal genotype. (d) Characterization of gametophytic maternal effects by test crosses. As the phenotype of developing or mature seeds is determined by the genotype of the female gametophyte, phenotypic segregation is observable in F1 progenies of test crosses. (e) Characterization of sporophytic maternal effects by test crosses. As the phenotype of developing or mature seeds is determined by the genotype of the female sporophyte, phenotypic segregation is unobservable in F1 progenies of test crosses.
It's worth noting that the tt mutants display another common phenotype of the non-pigmented seed coat[21]. However, regulation of such a seed phenotype is not attributed to canonical maternal effects because the integument-derived seed coat and its pigmentation are directly inherited from the mother generation (Fig. 2c). Likewise, maternal defects of the integument and megaspore are also not a consequence of canonical maternal effects because the filial generation is not involved as exemplified by the regulation conferred by AINTEGUMENTA (ANT)[22,23] and INNER NO OUTER (INO)[24].
In contrast, the integument influences on seed size are considered maternal effects[25]. The integument proliferation and expansion make the cavity for filial development. In this regard, the 'DA' pathway, where 'DA' means 'large' in Chinese, contains a group of genes in the ubiquitin pathway and regulates the seed size maternally and sporophytically[26,27]. TTG2[28], APETALA2 (AP2)[29], and ABERRANT TESTA SHAPE (ATS)[30,31] also act maternally to control integument characteristics. In addition, the signals from the maternal integument also control the outcome of filial development. Maternal siRNAs, which are produced in sporophytic tissues, such as the integument, or transcribed by the maternal alleles in the endosperm, repress the AGAMOUS-LIKE transcription factors (AGLs) in the endosperm to regulate endosperm development[32−34]. Moreover, several cytochrome P450s (CYPs), including KLUH (KLU; CYP78A5)[35], ENHANCER OF da1-1 3 (EOD3; CYP78A6)[36], and possibly CYP78A7/A9[36,37], generate mobile maternal signals to regulate seed size. Besides, maternal auxin provided by the integument regulates embryonic cell proliferation and patterning[38,39], while maternal gibberellin is crucial for the programmed cell death of the embryonic suspensor[40].
In monocots, there are several functionally conserved pathways that exert maternal effects. For example, the causal genes of the QTL GRAIN WEIGHT 2 (GW2) in rice[41], wheat[42], and maize[43] maternally regulate seed size. These genes encode E3 ligases homologous to DA2 in Arabidopsis. However, different seed structures of monocots and dicots (Fig. 1a) implicate partially distinct regulatory mechanisms. As the seeds of the grass family are usually merged with the pericarp and covered by husks, the maternal effects may be related to fruit or flower tissues. A lot of brassinosteroid-related mutants exhibit altered grain size and shape[44], which are at least attributed to the misregulation of cell division or expansion in lemma and palea. Besides, different seed structures also indicate different functional modes of homologous genes in monocots and dicots. For example, GIANT EMBRYO (GE) encodes CYP78A13 and regulates the balance of endosperm and embryo development in rice[45,46], while its Arabidopsis homologs do not exert such an effect.
In contrast to the father, both maternal sporophyte and female gametophyte are involved in the control of seed development. Thus, the maternal effects may act sporophytically or gametophytically. Maternal gametophytic effects refer to the phenotype of offspring determined by the haplotype inherited from the mother. It can be distinguished from the sporophytic effects by test crosses, where heterozygotes are used as maternal plants to be pollinated with the pollen from a homozygous donor (Fig. 2d). A maternal gametophytic effect results in a 1:1 segregation ratio in the progenies from the test cross, whereas a maternal sporophytic effect does not cause phenotypic segregation of the progenies from the test cross (Fig. 2e). Although the phenotype of the maternal gametophytic effect is similar to that caused by maternally imprinted genes (MEGs; see Fig. 3), they are conceptually different. A maternal gametophytic effect could be explained by the biologically active components inherited from the maternal gametophyte, while MEGs are responsible for de novo synthesis of biologically active components in the filial tissue. For example, ADRENODOXIN REDUCTASE (ADXR), ADRENODOXINS (ADXs), and their targets, mitochondrial cytochrome P450s, are important regulators of the mitochondrial steroidogenic pathway in female gametophytes, and they influence early embryogenesis in a maternal gametophytic manner[47].
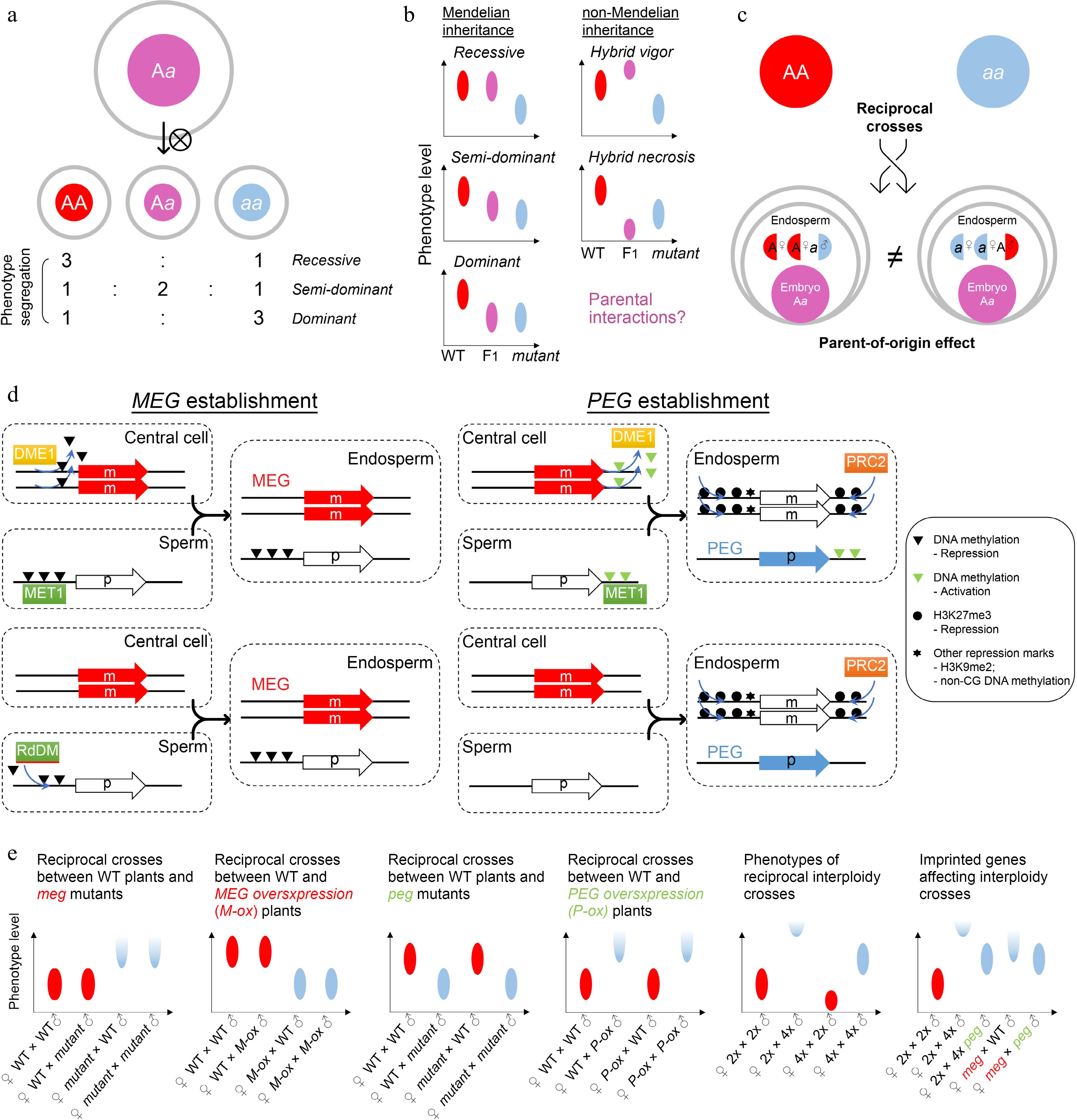
Figure 3.
Zygotic control of seed development. (a) A typical zygotic effect causes phenotype segregation among F1 siblings. (b) Scheme of Mendelian and non-Mendelian inheritance patterns of F1 progenies with zygotic effects. Left panels: possible phenotypes of F1 progenies with a recessive, semi-dominant, or dominant mutation. Right panels: possible phenotypes of F1 progenies with a non-mendelian mutation. Such patterns of non-mendelian inheritance are likely related to parental interactions. Ellipse indicates the quantified range of a phenotype. (c) Scheme of endospermic factors regulating F1 phenotypes in a parental-dependent manner. (d) Typical mechanisms underlying gene imprinting. MEG, maternally imprinted gene; PEG, paternally imprinted gene. (e) Phenotypic assumptions based on unbalanced parental dosage. The first four panels from the left show phenotypic patterns of reciprocal crosses between wild-type plants and plants with loss of function or overexpression of imprinted genes. The fifth panel shows phenotypic patterns of interploidy crosses in Arabidopsis, while the sixth panel shows a smilar phenotype between the paternal-excess cross (2nd column) and the cross with loss of MEG (meg) (4th column). Such phenotypes are suppressed by loss of PEG (peg) (3rd and 5th columns). Ellipse indicates the quantified range of a phenotype, while half ellipse indicates a possible abortive phenotype.
-
In contrast to maternal effects, zygotic effects delineate a phenomenon where the offspring phenotype is determined by its own genotype. Theoretically, any autonomous regulation of embryogenesis should show a zygotic effect. For example, the LEAFY COTYLEDON (LEC) class genes in Arabidopsis, including LEC1/2, ABSCISIC ACID INSENSITIVE3 (ABI3), and FUSCA3 (FUS3), are major regulators of embryogenesis and endosperm development[48−50]. The zygotic effects are manifested in these lec mutants as phenotypic segregation of individually developing seeds is observable in siliques of heterozygous plants (Fig. 3a). The filial LEC class genes are regulated by BABY BOOM (BBM)[51], which is one of the major inducers of early embryogenesis, and ectopic expression of BBM is sufficient to induce asexual and somatic embryo development[52,53]. As BBM is expressed in maternal sporophyte and gametophyte cells as well as filial zygotic cells[52], its effect on LEC genes implies a transition from maternal control to zygotic control. This transition is associated with the zygotic genome activation (ZGA)[54], in which both maternal and paternal genomes start to exert function in the filial cells.
In some special scenarios, filial phenotypes are superior or inferior to those of both parents, which is known as hybrid vigor (heterosis) or hybrid necrosis, respectively (Fig. 3b). These effects are not explained by the genetic background of F1 progenies, regardless of whether the mutation is recessive, dominant, or semi-dominant/dosage-dependent. Such patterns of non-Mendelian inheritance are likely related to parental interactions, including at the epigenetic level, although the mechanisms are so far unclear at the molecular level (Fig. 3b). Investigations so far have shown that hybrid necrosis is physiologically similar to auto-immunity and depends on the interactions between pairs of parent-of-origin compounds[55], while epigenetic regulation provides a possible platform for hybrid vigor[56,57] because the interaction of parental epi-alleles confers new characteristics in the F1 progenies. Overall, hybrid vigor and necrosis that influence the F1 seed development are special cases of zygotic effects[58], which are of particular value for crop breeding, including phenotypic improvement of filial generations[59−61].
Moreover, the endosperm is also considered as filial (zygotic) tissue. For canonically recessive or dominant mutants with endosperm defects, the filial segregation of heterozygous plants is observed as those with embryonic defects. For example, the maize ears of endosperm breakdown1 (enb1) heterozygous mutants contain normal or endosperm-defective kernels according to the genotypes of individual kernels[62]. However, whilst the genome is equally inherited from the parents (maternal : paternal = 1:1) in the embryo, the parental contributions are unequal in the endosperm (maternal : paternal = 2:1). Therefore, the F1 progenies of reciprocal crosses display different endosperm genotypes and parent-of-origin effects are thus expected in reciprocal crosses if the mutation is semi-dominant, dosage-dependent, or parentally biased (Fig. 3c). A group of genes with allele-specific expression depending on their parental origin is called imprinted genes. These genes are known as MEGs or paternally expressed genes (PEGs), as their transcription is only activated in the allele inherited from the mother or father, respectively. Imprinted genes are mainly found in the endosperm of both monocots and dicots[63−66] and regulate seed phenotypes in a parent-of-origin manner (Fig. 3c). The establishment of imprinting generally requires high activity of DNA demethylation and H3K27me3 deposition in the central cell as well as early endosperm, compared to the sperm[67].
The establishment of MEGs can be achieved by relieving MEGs from repression compared to the paternal allele, which depends on the passive activation of DEMETER (DME; eraser for DNA methylation) in the central cell[68] (Fig. 3d). Consequently, loss of maternal DME leads to seed abortion because of impaired imprinting[69]. Meanwhile, mutation of METHYLTRANSFERASE 1 (MET1), which is a CG DNA methyltransferase, also leads to a parent-of-origin effect on seed size because of the imprinting disturbance in the endosperm[70]. The biased parental DNA methylation by DME and MET1 accounts for the maternal allele-specific expression of FLOWERING WAGENINGEN (FWA), FERTILIZATION INDEPENDENT SEED 2 (FIS2), and MATERNALLY EXPRESSED PAB C-TERMINAL (MPC)[71−73], and at least partially for MEDEA (MEA)[69,72,74,75]. Besides, the paternal alleles of some MEGs can be actively silenced by the noncanonical RNA-directed DNA methylation (RdDM) pathway activated in nurse cells of gametes[76−79], which leaves the remaining maternal allele active in the endosperm (Fig. 3d).
However, except for the parental differential DNA methylation, a significant number of MEGs are established by unknown mechanisms beyond DNA methylation[80,81]. As the putative MEGs could have been contaminated by the genes expressed in maternal sporophytic tissues[82], it remains an open question if alternative pathways for MEG establishment exist or not. In addition, some imprinted genes are not conserved among different accessions of the same species[80], suggesting that the imprinting status could be dynamic or altered during evolution.
The establishment of PEGs is achieved mainly by silencing of the maternal allele (Fig. 3d). For example, it is hypothesized that DNA methylation of the 3' flanking sequence of PHERES 1 (PHE1), a PEG, excludes H3K27me3 deposition in the endosperm. The maternal allele of PHE1 is demethylated in the central cell by DME, facilitating subsequent H3K27me3-mediated silencing in the endosperm, whereas the paternal allele of PHE1 keeps DNA methylation, which is maintained by MET1 and remains active in the endosperm[83,84]. As an AGAMOUS-LIKE transcription factor, PHE1 further controls the imprinting of other loci in the endosperm[85]. In a genomic view, some PEGs are downregulated when the paternal loss of MET1 is introduced[86], indicating that paternal DNA methylation is common for PEG activation. Although H3K27me3 is a core silencing mark for the maternal alleles of PEGs[87], these loci are not highly correlated with DME-mediated DNA demethylation[80]. It is possible that H3K27me3 itself functions to build the parental asymmetry independently of DNA methylation (Fig. 3d). Notably, silencing of the maternal allele of PEGs by H3K27me3 is frequently associated with non-CG DNA methylation and H3K9me2 histone modifications, which could be the subsequent mechanisms contributing to imprinting establishment[88]. Given that the maternal PRC2 components, such as MEA and FIS2, regulate PEGs as well, maternal regulation is generally more dominant over paternal regulation[89]. This is in agreement with the notion that seed development relies on maternal tissues.
Although many imprinting genes have been identified, most mutants of imprinted genes (especially PEGs) do not show obvious phenotypes[90,91] except that there are increasing literature reporting the link between imprinted genes and potential seed phenotypes[92,93]. The most known imprinting-related phenotype is seed abortion caused by endosperm overproliferation and cellularization failure. Theoretically, a seed phenotype related to a given imprinted gene is either maternally or paternally determined, although imprinted genes fundamentally function zygotically (Fig. 3e). Interestingly, a dramatic parent-of-origin effect is observed in the interploidy reciprocal cross in Arabidopsis, where the F1 progenies from the reciprocal parental origins show opposite phenotypes[94,95]. Similar phenomenon in reciprocal interploidy crosses is also reported in monocots, such as maize and rice[96−98]. Although tetraploid seeds are generally larger than diploid seeds, the F1 seeds from the maternal excess cross (♀tetraploid × diploid♂) are smaller than diploid seeds (precocious endosperm cellularization), while the F1 seeds from the paternal excess cross (♀diploid × tetraploid♂) are larger than tetraploid seeds or even aborted (delayed/failed endosperm cellularization) (Fig. 3e). Like the hybrid vigor or necrosis, such patterns of non-Mendelian inheritance in interploidy crosses indicate parental interactions.
Opposite phenotypes between maternal excess and paternal excess crosses imply that MEGs and PEGs tend to restrict and promote endosperm growth, respectively. This is also supported by the findings that seed abortion caused by defective MEGs can be partially rescued by additional loss of some PEGs in Arabidopsis[84,99]. In particular, PEGs are critical for seed abortion caused by paternal excess, which is known as the triploid block or interploidy barrier. Mutants of several PEGs and mutants with failed PEG establishment suppress the phenotype of paternal excess[77,90,99−105], although they do not cause visible defects per se in diploids (Fig. 3e). Global paternal demethylation bypasses triploid block[106,107], suggesting that paternal epigenome is crucial. It is possible that imprinted genes function together as a genomic feature rather than acting as individual regulators to regulate seed phenotypes. Therefore, obvious seed phenotypes are only found in the genetic backgrounds with dramatic or global disturbance of parental balance, such as interploidy progenies or mutants of imprinted genes that are epigenetic mark builders and general transcription factors.
-
Different cell types with distinct genetic backgrounds are involved in seed development, and the communications among cell types make the underlying regulations more complicated and cannot be simply interpreted as maternal or zygotic effect. Signal communications among multiple cell types are expected, but the relevant mechanisms are largely obscure. For example, pollen-delivered SHORT SUSPENSOR (SSP) mRNA is only translated in the zygote soon after fertilization to regulate the zygotic YODA (YDA) pathway, thereby controlling the asymmetric division[108,109], while paternal OsBBM1 transcripts delivered by the pollen trigger early embryogenesis in rice[110,111]. In addition, paternal miR159 represses central cell-inherited MYB33/65 to allow endosperm nuclear division[112] (Fig. 4a). Such paternal triggers could be more widespread, as there is evidence showing that pollen tube contents can mimic fertilization and induce the growth of maternal sporophytic tissues[113].
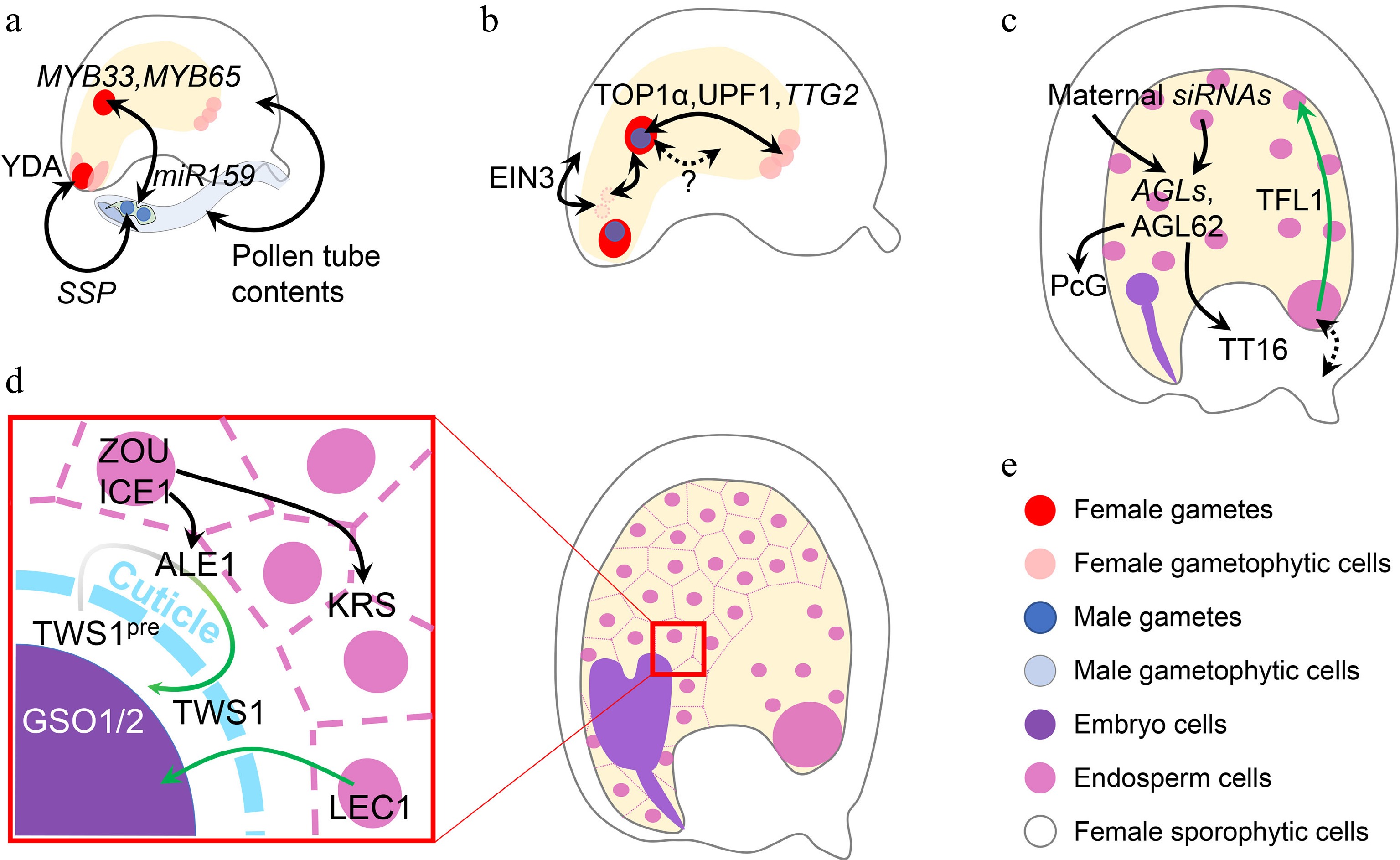
Figure 4.
Inter-tissue communication in seed development. (a) Inter-tissue communication at the beginning of seed development. Paternal SSP mRNA from the pollen affects the zygotic YDA pathway to determine zygote division. Paternal miR159 from the pollen quenches maternal MYB33/65 to initiate nascent endosperm division. Other contents in the pollen tube can also trigger ovule growth, which mimicks fertilization. (b) Inter-tissue communication in early seed development. Synergid nuclei affect the maternal-paternal genome ratio in the nascent endosperm, which is oppositely regulated by sporophytic and gametophytic EIN3. The communication between maternal antipodal cells and paternal cues in the nascent endosperm relies on the relative dosage of maternal and paternal TTG2, which is transcriptionally regulated by TOP1α and UPF1. Nascent endosperm-female gametophyte communication is also suggested, although the mechanisms are yet unknown. (c) Inter-tissue communication regulating endosperm and integument development. Chalazal-transcribed TFL1 functions in the peripheral endosperm to regulate endosperm cellularization. This module also infers a potential maternal-filial communication at the chalazal part. Endosperm regulators, AGLs, are regulated by maternal siRNAs from both the endosperm and maternal tissues. AGL62 in turn regulates maternal nucellus degradation via the maternal TT16 and integument growth via the maternal PcG complex. (d) Inter-tissue communication between endosperm and embryo. When the cuticle barrier between endosperm and embryo is not established, endosperm-expressed LEC1 relocates into the embryo to exert its function. The integrity of such a barrier is monitored by two-way communication, in which the precursor of the embryo-expressed TWS1 peptide (TWS1pre) is processed in the endosperm by ALE1 and the mature peptide signal moves back into the embryo to activate the GSO1/2-pathway. (e) Color legend shows different elements in this figure. Single- and double-headed arrows indicate one-way and reciprocal regulations, respectively. Dashed arrow indicates a putative regulation. Green single-headed arrow indicates protein movement, while green gradient single-headed arrow indicates protein movement along with the maturation process.
Parental communications are crucial at the initial stage of seed development, especially with regard to the female gametophytic cues and nascent endosperm (Fig. 4b). It has been recently revealed that nascent endosperm growth is related to the disintegration of synergid-derived nuclei, which affects the overall maternal-paternal ratio in the nascent endosperm. The disintegration of synergid nuclei is inhibited by maternal sporophytic ETHYLENE INSENSITIVE 3 (EIN3), but promoted by gametophytic EIN3[114]. Besides, the nitrous oxide treatment in maize causes defective endosperms in the affected kernel, where the dosage balance between the newly synthesized compounds in nascent endosperm and compounds inherited from female gametophytes is changed without affecting the maternal-paternal balance in the endosperm, implying an as-yet-unknown female gametophyte-endosperm communication[115,116]. Such communication is also found in Arabidopsis. In addition to the well-known sporophytic functions of TTG2, the relative parental dosage of gametophytic TTG2 also affects the final seed size[117]. Maternal TTG2 is expressed in antipodal cells, while paternal TTG2 is expressed in the nascent endosperm inherited from the sperm. Such a parental module influences the outcome of interploidy crosses, although it is still unknown why the maternal and paternal TTG2 exert antagonistic functions[117]. Interestingly, antipodal cells, whose foci are later replaced by chalazal endosperm, are most extensively regulated by parental cues[118]. The chalazal part is also crucial for nutrient exchange between maternal and filial tissues[119], echoing the principles inferred by the parental conflict hypothesis[120]. These findings hint that it may be common for a gene to act oppositely in different parental contexts at the beginning of seed development.
At the late seed development stage, TERMINAL FLOWER1 (TFL1) regulates seed size by affecting endosperm cellularization. TFL1 is transcribed in the chalazal endosperm, but its protein is relocated into the peripheral endosperm to take action (Fig. 4c). Genetic data reveals the maternal effect of TFL1 mutation, suggesting a potential signal exchange between maternal and zygotic tissues[121]. At this stage, endosperm-maternal sporophyte communications are common. The major endosperm regulator AGAMOUS-LIKE 62 (AGL62)[122], which is not imprinted but under the paternal control of PHE1[85], can guide maternal nucellus degradation by promoting maternal TT16 expression[123], and regulates auxin transport from the endosperm to the integument to repress maternal Polycomb Group (PcG) function[124] (Fig. 4c). Considering the previously mentioned maternal regulation of the endosperm AGLs by siRNAs[32−34], reciprocal ways exist for the paternal-maternal antagonism. Although more details remain to be revealed regarding the maternal-paternal interaction, the paternal effect is generally weaker than the maternal one[125].
After endosperm cellularization, the endosperm-embryo communications are crucial for the seed maturation process (Fig. 4d). The endosperm-produced LEC1 is transported into the embryo to participate in the transcriptional programming during embryo maturation[49]. The overall LEC1 expression level is more correlated to the maternal allele because of the 2:1 genome ratio in the endosperm. Meanwhile, an embryo-produced peptide TWISTED SEED1 (TWS1) is processed by endosperm-expressed subtilisin-like protease ABNORMAL LEAF-SHAPE 1 (ALE1) and in turn perceived by embryo-presented receptors GASSHO1/2 (GSO1/2) and their co-receptors[126,127] when the cuticle barrier between embryo and endosperm is not fully established. This two-way communication acts downstream of the regulatory module in which the endosperm-expressed transcription factors ZHOUPI (ZOU) and INDUCER OF CBF EXPRESSIONICE 1 (ICE1) regulate embryo development by controlling the expression of ALE1 in the embryo-surrounding region (ESR)[128,129] and another putative signal-function peptide KERBEROS (KRS) in the endosperm[130]. These components function together to build a molecular sensor for cuticle integrity between endosperm and embryo, which is a marker delineating seed development stages.
-
Apart from the extensive studies showing the nature of parental regulation on seed development in Arabidopsis, emerging studies have also shown that such regulations are valuable for crop engineering. In rice, a significant number of imprinted genes are associated with grain yield quantitative trait loci with the potential function of regulating nutrient metabolism and endosperm development[131]. These findings echo the parental conflict hypothesis: mothers restrict the resource allocation for seed development to feed all their descendants, while fathers help their offspring evade this maternal restriction[120]. Therefore, investigation of parental regulations on seed development is certainly important for improving seed yield and quality for various crops.
Because of the tissue complexity and genetic diversity, histological and genetic analyses are essential for functional studies of potential parental interactions. However, such data only provide a rough framework to assess the nature of a potential regulation, while the detailed mechanisms must be revealed by other combined approaches. Traditional biochemical assays and in vitro tests have inherent disadvantages in revealing mechanisms during seed development because of missing of intercellular information, which is, however, critical for understanding seed development. Emerging single-cell technologies are likely good platforms to reveal cellular relationships during seed development. Using single-cell technologies, DNA methylation, chromatin accessibility, protein abundance, and gene perturbation can be investigated at the sub-tissue level[132]. For example, single-nucleus sequencing of Arabidopsis endosperm has revealed the functional partitioning among endosperm nuclei, with the chalazal endosperm showing the most parentally biased expression[118]. This is consistent with the fact that the chalazal part is the interface of maternal and filial tissues, which could be the frontline of maternal-filial signal communications. With a clear understanding of the parental interplay among various cell types involved in seed development, valuable molecular targets could be identified and precisely modified for crop improvement.
This work was supported by Singapore Food Story R&D Programme (SFS_RND_SUFP_001_04) and the Reimagine Research Grant from the National University of Singapore. We apologize to authors whose excellent work could not be cited owing to space limitations.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2022 by the author(s). Published by Maximum Academic Press on behalf of Hainan Yazhou Bay Seed Laboratory. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Li C, Yu H. 2022. Parental regulation of seed development. Seed Biology 1:7 doi: 10.48130/SeedBio-2022-0007
Parental regulation of seed development
- Received: 23 October 2022
- Accepted: 17 November 2022
- Published online: 07 December 2022
Abstract: Angiosperms produce seeds with two zygotic tissues, namely the embryo and endosperm, from a unique double fertilization process. Seed development occurs within the maternal tissue and relies on maternal resources. Paternal tissue is not directly involved in seed development, and paternal regulation is usually based on the paternal genome of zygotic tissues in the filial generation. The complicated maternal-paternal communications and maternal-zygotic interactions result in distinguishable genetic effects on seed development. Here we review the conceptual framework of parental regulations on seed development. We summarize the common seed development process and look into the regulations pertaining to maternal and zygotic effects. Examples with more complicated interactions at the inter-tissue level are also discussed in the context of interwoven parental regulations.
-
Key words:
- embryo /
- seed development /
- Parental regulation /
- maternal effect /
- zygotic effect /
- endosperm













