-
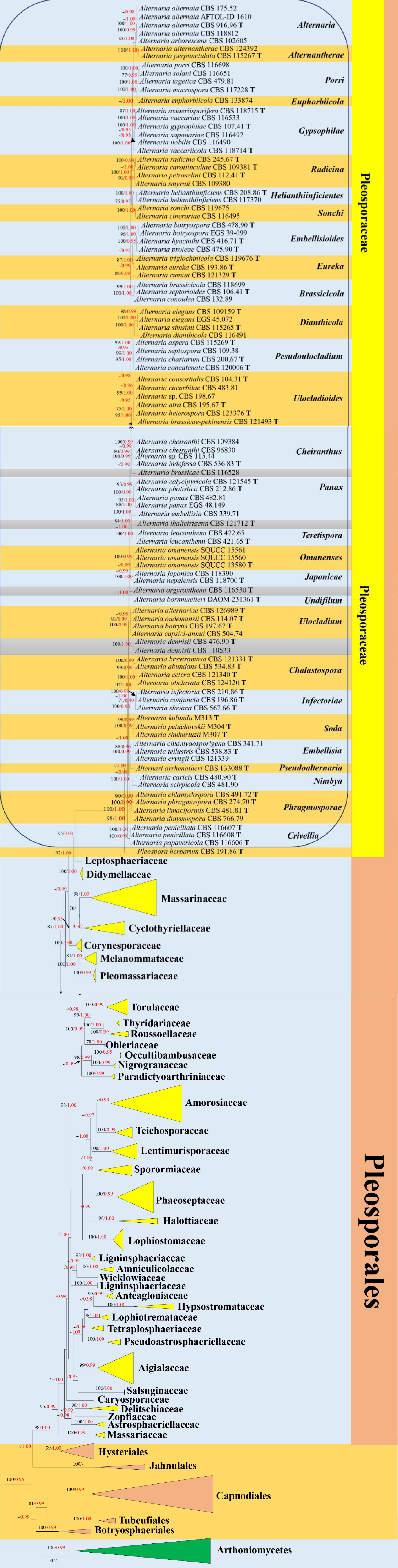
Figure 1.
Phylogenetic construction of Pleosporales using RAxML-based maximum likelihood analysis of a combined ITS, LSU, SSU, TEF1-α and RPB2 DNA sequence dataset. Bootstrap support values for maximum likelihood (ML, black) equal to or greater than 70% and Bayesian posterior probabilities (PP, red) equal to or greater than 0.95 PP are shown above the nodes. The tree is rooted to Arthoniomycetes (Arthonia dispersa UPSC2583, Dendrographa leucophaea f-minor, Roccella fuciformis Tehler 8171 and Schismatomma decolorans Ertz 5003). The type strains are indicated by boldface 'T'.
-
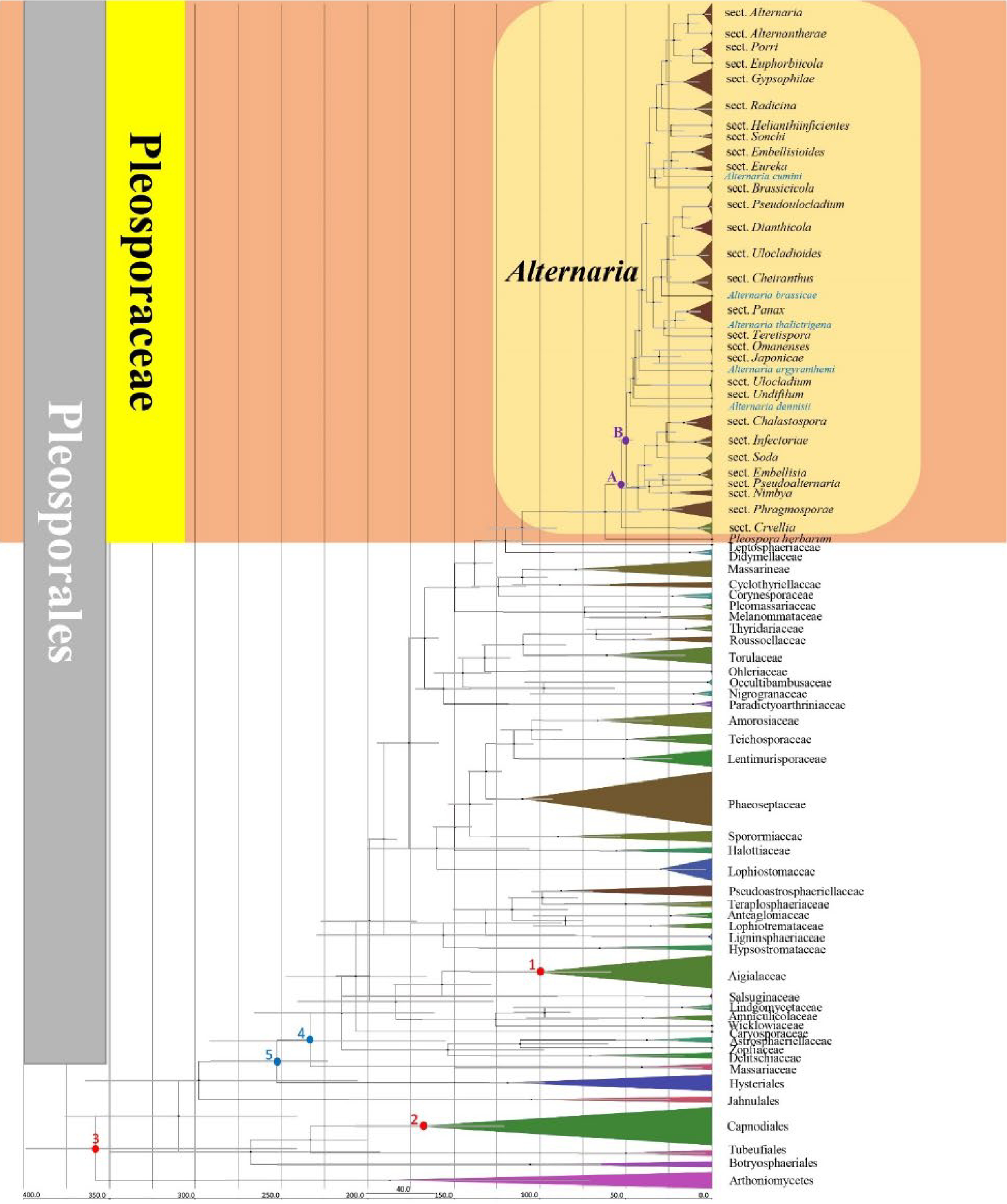
Figure 2.
Maximum clade credibility (MCC) tree with divergence times estimates obtained from BEAST. Numbers in red indicate the fossil (1, 2) and secondary (3) points. Numbers in blue indicate divergence time estimate of Pleosporales (stem age: 5, crown age: 4). Letters in purple indicate divergence time estimate of Alternaria (stem age: A, crown age: B). Single lineages of Alternaria species are highlighted in blue.
-

Figure 3.
Phylogenetic construction of genus Alternaria using RAxML-based maximum likelihood analysis of a combined ITS, LSU, SSU, TEF1-α, RPB2, GAPDH and Alt-a1 DNA sequence dataset. Bootstrap support values for maximum likelihood (ML, black) equal to or greater than 70% and Bayesian posterior probabilities (PP, red) equal to or greater than 0.95 PP are shown above the nodes. The tree is rooted to Stemphylium vesicarium (CBS 191.86) and Pleospora tarda (CBS 714.68). Newly generated strains are in blue. The type strains obtained from ex-type cultures are indicated by 'T' and the type strains obtained from specimens are indicated by 'H'.
-
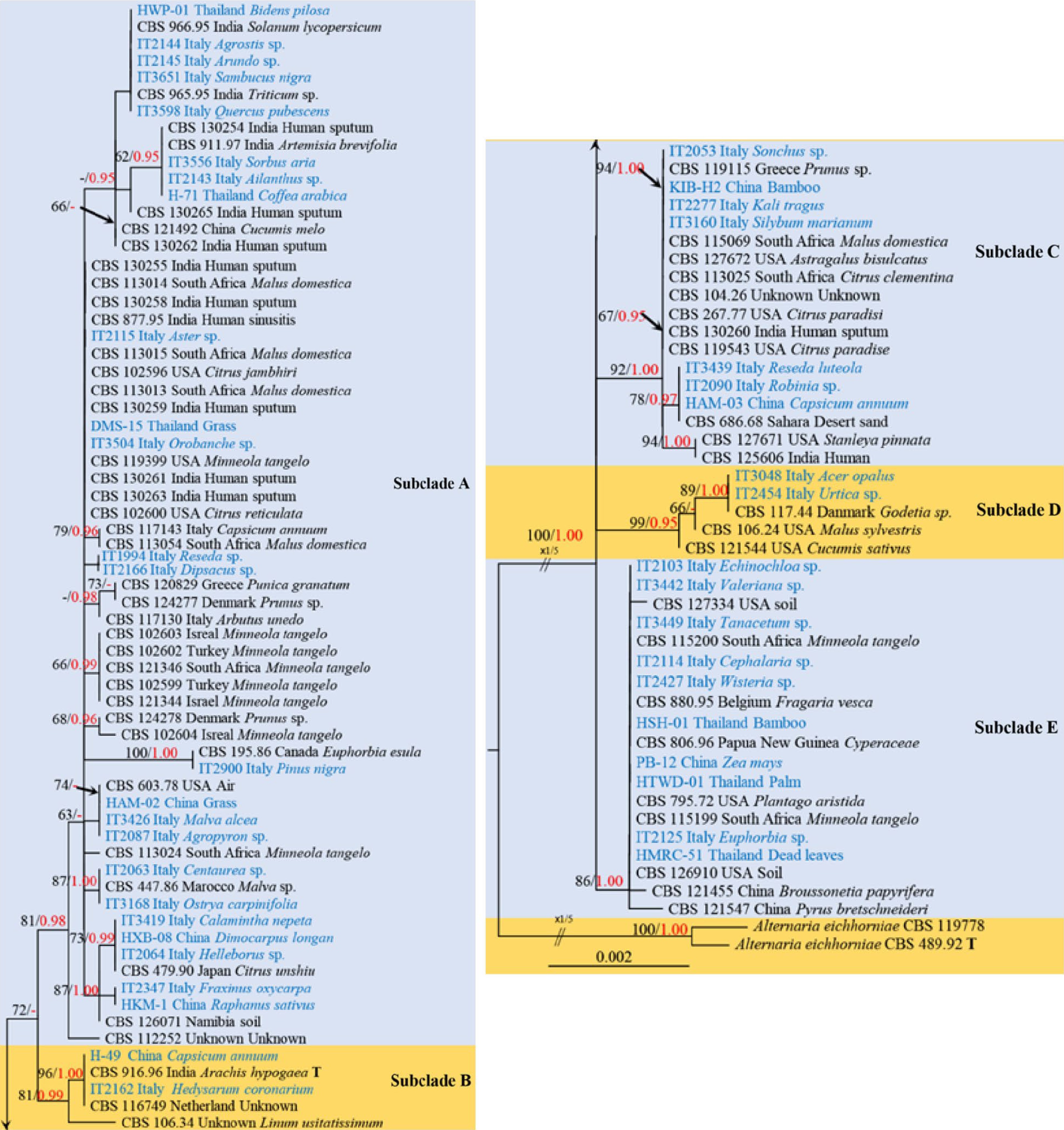
Figure 4.
Phylogenetic construction of Alternaria alternata using RAxML-based analysis of a combined ITS, LSU, SSU, TEF1-α, RPB2, GAPDH and Alt-a1 DNA sequence dataset. Bootstrap support values for maximum likelihood (ML, black) equal to or greater than 60% and Bayesian posterior probabilities (PP, red) equal to or greater than 0.95 are shown above the nodes. The tree is rooted to Alternaria eichhorniae (CBS 489.92) and Alternaria eichhorniae (CBS 119778). Newly generated strains are in blue, and the type strains are indicated by 'T'.
-

Figure 5.
Phylogenetic construction of Alternaria sect. Infectoriae using RAxML-based analysis of a combined ITS, GAPDH and ATPase DNA sequence dataset. Bootstrap support values for maximum likelihood (ML, black) equal to or greater than 70% and Bayesian posterior probabilities (PP, red) equal to or greater than 0.95 PP are shown above the nodes. The tree is rooted with taxa in sect. Chalastospora (Alternaria malorum CBS 135.31 and A. abundans CBS 534.83). Newly generated strains are in blue. The type strains obtained from ex-type cultures are indicated by 'T' and the type strains obtained from specimens are indicated by 'H'.
-
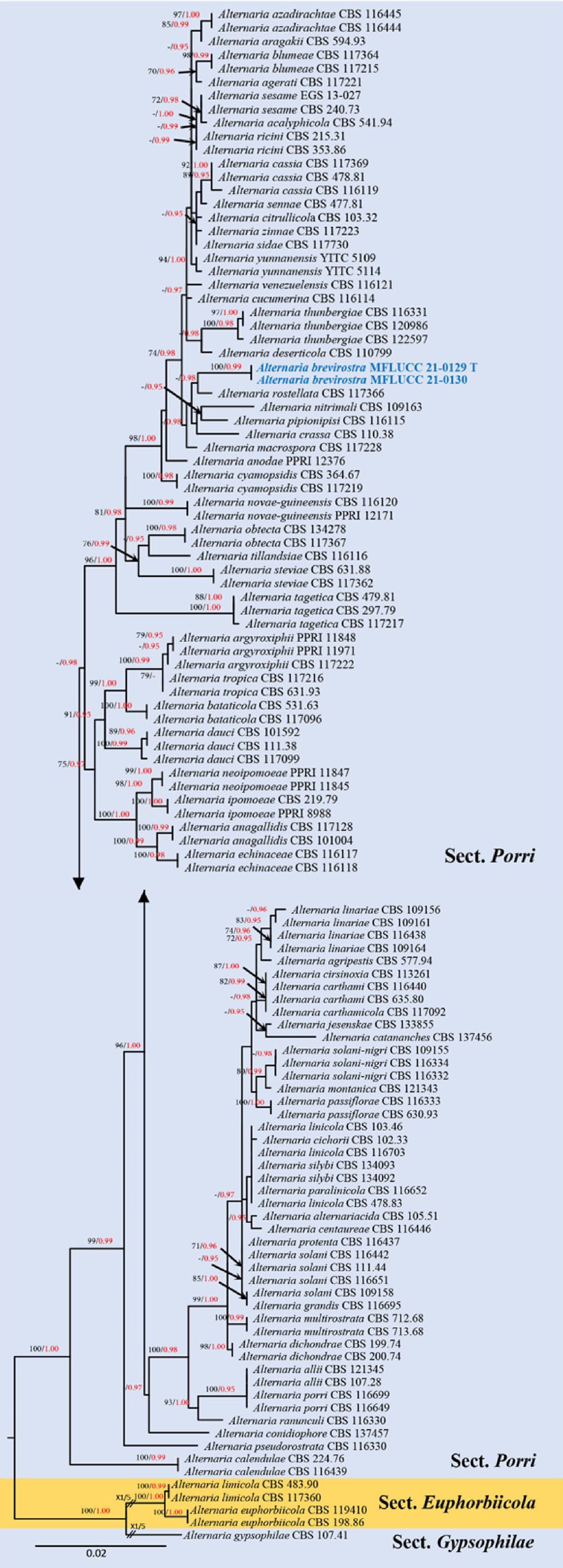
Figure 6.
Phylogenetic construction of Alternaria sect. Porri using RAxML-based analysis of a combined ITS, GAPDH, TEF1-α, RPB2 and Alt-a1 DNA sequence dataset. Bootstrap support values for maximum likelihood (ML, black) equal to or greater than 70% and Bayesian posterior probabilities (PP, red) equal to or greater than 0.95 PP are shown at the nodes. The tree is rooted to sect. Gypsophilae (Alternaria gypsophilae CBS 107.41). Newly generated strains are in blue and the type strains are indicated by 'T'.
-
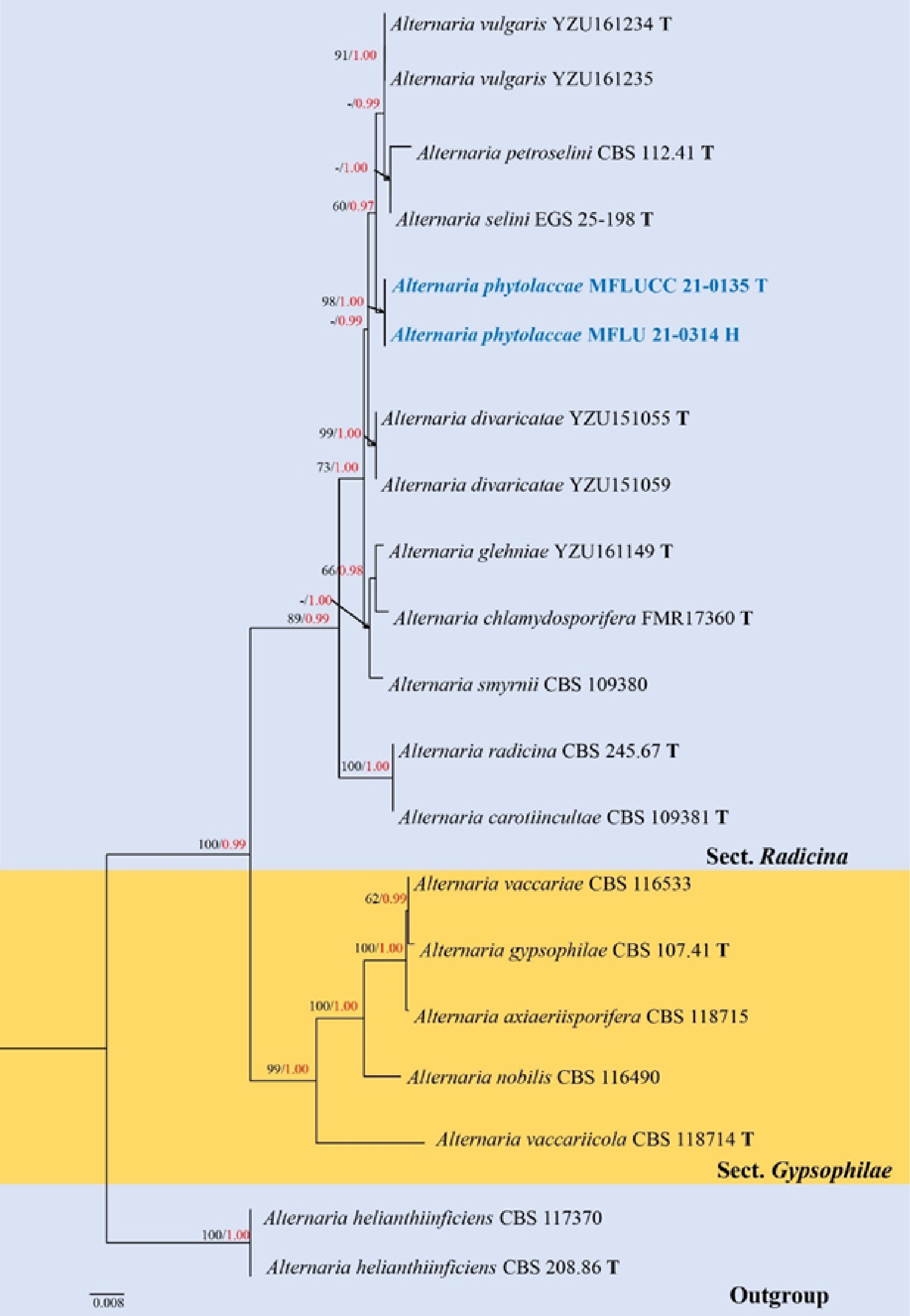
Figure 7.
Phylogenetic construction of Alternaria sect. Radicina using RAxML-based analysis of a combined ITS, TEF1-α, GAPDH and Alt-a1 DNA sequence dataset. Bootstrap support values for maximum likelihood (ML, black) equal to or greater than 60% and Bayesian posterior probabilities (PP, red) equal to or greater than 0.95 PP are shown above the nodes. The tree is rooted to A. helianthiinficiens (CBS 208.86 and CBS 117370). Newly generated strains are in blue. The type strains obtained from ex-type culture are indicated by 'T' and the type strains obtained from holotype specimen are indicated by 'H'.
-

Figure 8.
Sporulation in Alternaria spp. (a) A. eupatoriicola (MFLU 21-0319). (b) A. lathyri (MFLU 21-0297). (c) A. oblongoellipsoidea (MFLU 21-0310). (d) A. macilenta (MFLU 21-0305). (e) A. baoshanensis (MFLU 21-0296). (f) A. falcata (MFLU 21-0306). (g) A. ellipsoidialis (MFLU 21-0307). (h) A. arctoseptata. (i) A. salicicola (MFLU 21-0320). (j) A. arundinis (MFLU 21-0313). (k) A. phytolaccae (MFLU 21-0314). (l) A. breviconidiophora (MFLU 21-0317). (m) A. phragmiticola (MFLU 21-0316). (n) A. minimispora (MFLU 21-0295). Scale bars: (a)–(n) = 10 µm.
-

Figure 9.
Alternaria alternata (MFLU 21-0302). (a), (b) Colonies on dead branch. (c)–(i) Conidiophores. (j)–(u) Conidia. Scale bars: (a) = 0.1 cm, (b) = 300 µm, (c)–(u) = 20 µm.
-

Figure 10.
Alternaria arctoseptata (MFLU 21-0308, holotype). (a) Colonies on dead stem of Lathyrus sp. (Fabaceae). (b)–(g) Conidiophores bearing conidiogenous cells. (h), (i) Immature conidia. (j), (o) Conidia formed secondary conidiophores. (k)–(n) Mature conidia. Scale bars: (a) = 200 µm, (b)–(o) = 20 µm.
-

Figure 11.
Alternaria baoshanensis (MFLU 21-0296, holotype). (a) Colonies on dead rattan of Curcubita moschata. (b)–(e) Conidiophores with a distinct conidiogenous locus. (f)–(l) Variation in shape of conidia. Scale bars: (a) = 200 µm, (b)–(e) = 30 µm, (f)–(l) = 20 µm.
-

Figure 12.
Alternaria breviconidiophora (MFLU 21-0317, holotype). (a) Colonies on dead branch. (b) Conidiophores arising on stomatic base. (c)–(g) Conidiophores. (h)–(q) Conidia. Scale bars: (a) = 50 µm, (b)–(g) = 20 µm, (h)–(q) = 10 µm.
-

Figure 13.
Alternaria doliconidium (MFLU 21-0294). (a) Dead stem of Reseda sp. (Resdaceae). (b) Colonies on dead stem. (c)–(j) Conidiophores on natural substrate. (k)–(m) Conidiogenesis sporulated in vitro. (n) Germinated conidium. (o)–(r) Conidia ((q), (r) on natural substrate)). Scale bars: (a) = 0.1 cm, (b) = 500 µm, (c)–(r) = 20 µm.
-

Figure 14.
Alternaria ellipsoidialis (MFLU 21-0307, holotype). (a) Colonies on dead hulls of Brassica sp. (b)–(g) Conidiophores bearing conidiogenous cells with a few apical conidiogenous loci. (h)–(q) Conidia. Scale bars: (a) = 300 µm, (b)–(g) = 30 µm, (h)–(q) = 20 µm.
-

Figure 15.
Alternaria eupatoriicola (MFLU 21-0319, holotype). (a) Colonies on dead hanging stem of Eupatorium cannabinum. (b)–(f) Conidiophores bearing conidiogenous cells. (g)–(q) Conidia. Scale bars: (a) = 300 µm, (b)–(m) = 20 µm.
-

Figure 16.
Alternaria falcata (MFLU 21-0306, holotype). (a) Colonies on dead hanging stem of Atriplex sp. (b)–(g) Conidiophores bearing conidiogenous cells. (h)–(m) Conidia. Scale bars: (a) = 150 µm, (b)–(m) = 20 µm.
-

Figure 17.
Alternaria lathyri (MFLU 21-0297, holotype). (a) Colonies on dead stem dead aerial stem of Lathyrus sp. (b) Conidiophores and conidia. (c)–(h) Conidiophores bearing conidiogenous cells. (i)–(n) Conidia. (o) Germinated conidium. Scale bars: (a) = 200 µm, (b)–(o) = 20 µm.
-
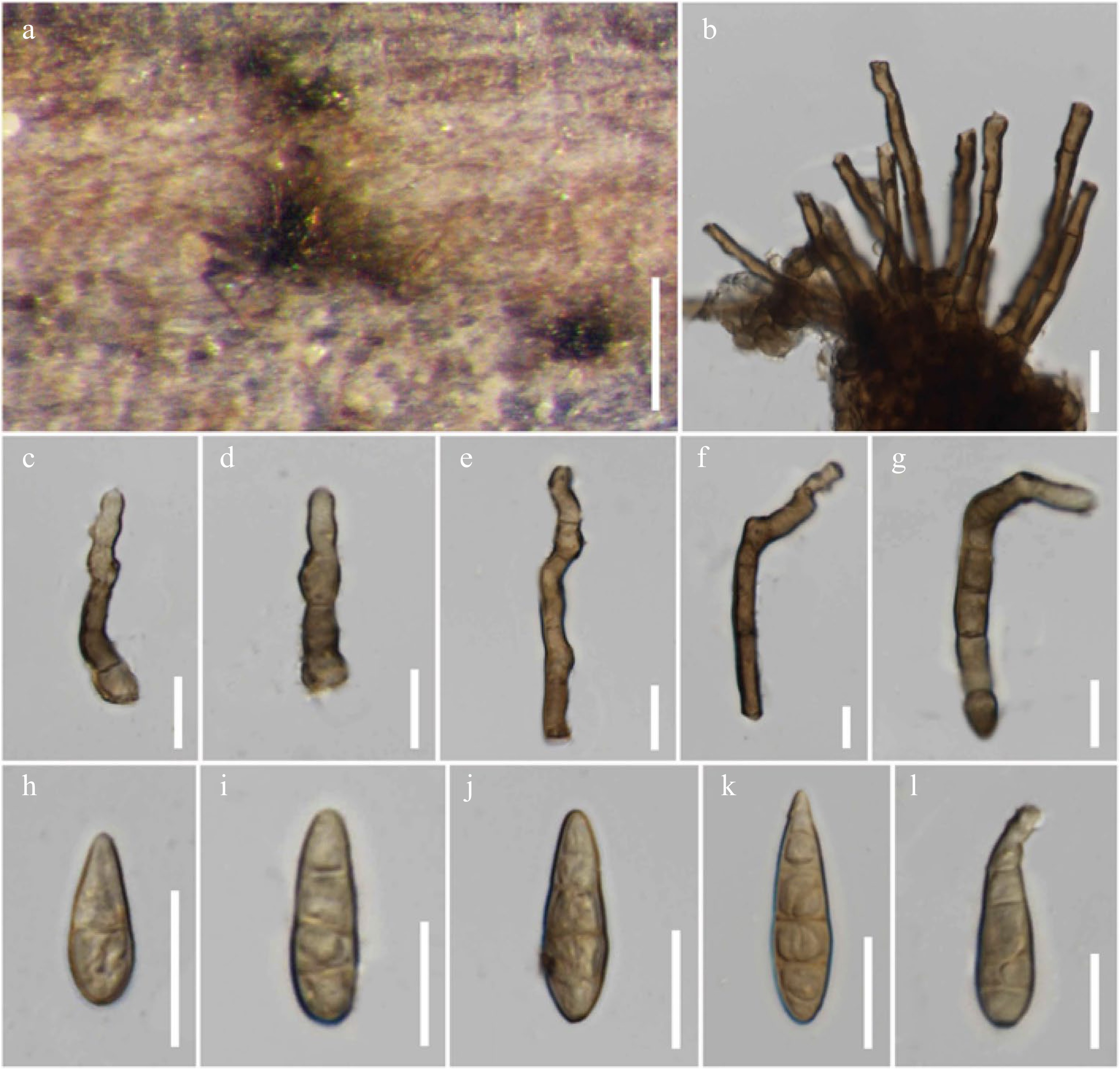
Figure 18.
Alternaria macilenta (MFLU 21-0305, holotype). (a) Colonies on dead stem of Scabiosa sp. (b)–(g) Conidiophores. (h)–(l) Conidia. Scale bars: (a) = 150 µm, (b)–(l) = 20 µm.
-

Figure 19.
Alternaria macroconidia (MFLU 21-0301, holotype). (a) Specimen examined of Spartium junceum (Fabaceae). (b) Colonies on dead aerial branch of Spartium junceum. (c)–(j) Conidiophores bearing conidiogenous cells. (k)–(n) Immature conidia. (o)–(s) Mature conidia. (t) Germinated conidium. Scale bars: (a) = 0.5 cm, (b) = 1000 µm, (c)–(t) = 20 µm.
-

Figure 20.
Alternaria minimispora (MFLU 21-0295, holotype). (a) Colonies on rotten peel of Citrullus lanatus. (b)–(e), (h), (i) Conidiophores bearing conidiogenous cells. (f) Conidiogenous cells. (g) Secondary conidiophores arising from conidia. (j)–(q) Conidia. Scale bars: (a) = 300 µm, (b)–(e), (h), (i) = 30 µm, (f), (g), (j)–(q) = 10 µm.
-

Figure 21.
Alternaria oblongoellipsoidea (MFLU 21-0310, holotype). (a) Colonies on dead stem of Cichorium intybus. (b)–(h) Conidiophores bearing conidiogenous cells. (i)–(l) Conidia. (m) Germinated conidium. Scale bars: (a) = 300 µm, (b)–(m) = 20 µm.
-

Figure 22.
Alternaria orobanches (MFLU 21-0303, holotype). (a) Colonies on dead stem of Orobanche sp. (b)–(g) Conidiophores bearing conidiogenous cells. (h) Conidiophores bearing conidiogenous cells with attached conidia. (i)–(q) Conidia. (r) Germinated conidium. Scale bars: (a) = 100 µm, (b)–(r) = 20 µm.
-

Figure 23.
Alternaria phragmiticola (MFLU 21-0316, holotype). (a) Colonies on dead stem Phragmites sp. (b)–(g) Conidiophores bearing conidiogenous cells. (h)–(o) Conidia. Scale bars: (a) = 200 µm, (b)–(o) = 20 µm.
-

Figure 24.
Alternaria salicicola (MFLU 21-0320, holotype). (a) Colonies on dead twig of Salix alba. (b)–(e) Conidiophores bearing conidiogenous cells. (f)–(p) Conidia. (q) Germinated conidium. Scale bars: (a) = 150 µm, (b)–(q) = 20 µm.
-

Figure 25.
Alternaria arundinis (MFLU 21-0313, holotype). (a) Colonies on dead stem of Arundo donax. (b)–(f) Conidiophores bearing conidiogenous cells. (g)–(i) Conidia formed secondary conidiophores. (j)–(n) Conidia. Scale bars: (a) = 150 µm, (b)–(n) = 20 µm.
-

Figure 26.
Alternaria nodulariconidiophora (MFLU 21-0315, holotype). (a) Colonies on dead stem of Heracleum sphondylium. (b)–(e) Geniculate conidiophores bearing conidiogenous cells. (f)–(m) Conidia. (n) Germinated conidium. Scale bars: (a) = 100 µm, (b)–(n) = 20 µm.
-
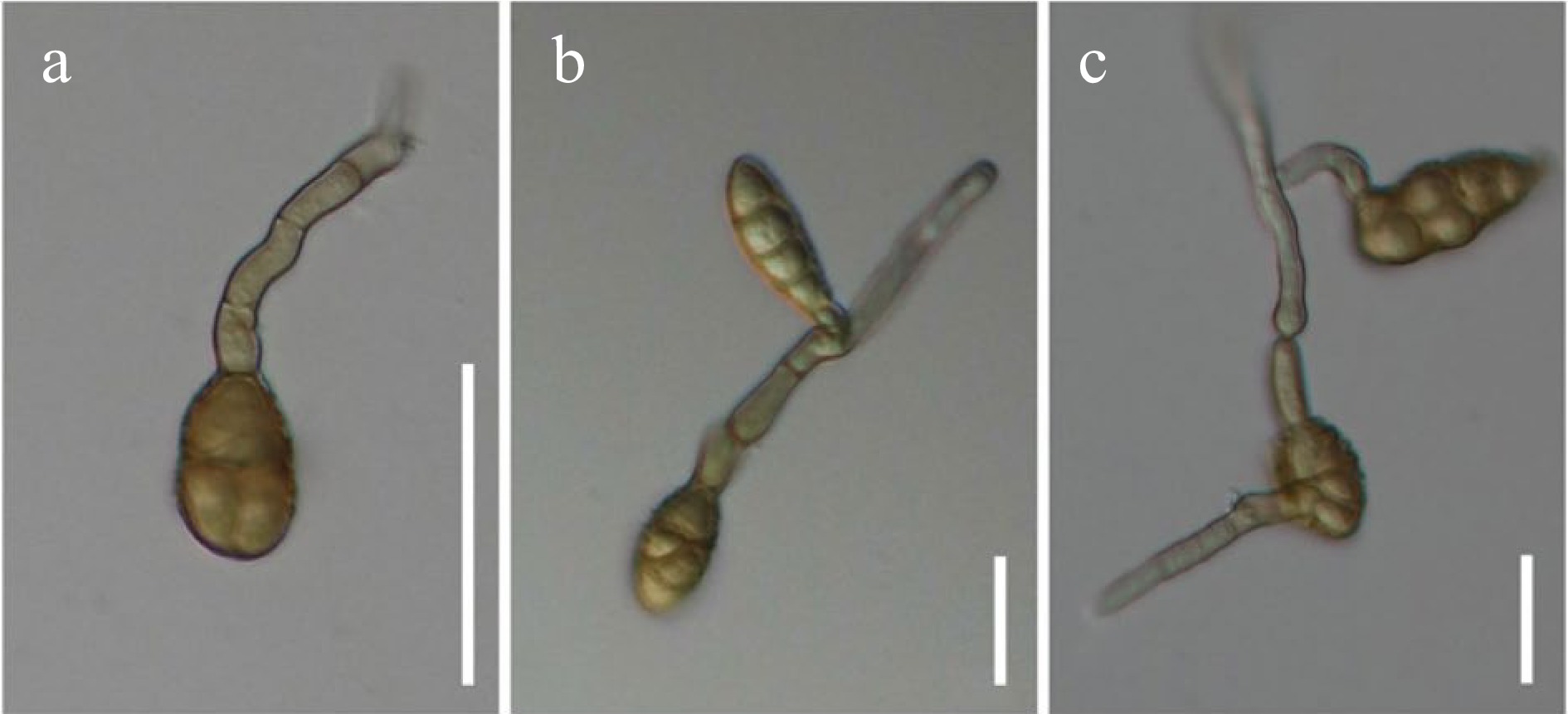
Figure 27.
Alternaria nodulariconidiophora (MFLUCC 21-0131) sporulated on PDA. (a) Conidium formed a geniculate secondary conidiophore. (b), (c) Secondary conidiophores bearing conidia with three-dimensional branching patterns. Scale bars: (a)–(c) = 30 µm.
-

Figure 28.
Alternaria brevirostra (MFLU 21-0312, holotype). (a) Colonies on dead stem of Plantago sp. (b)–(f) Conidiophores bearing conidiogenous cells. (g)–(k) Conidia. (l) Germinated conidium. Scale bars: (a) = 100 µm, (b)–(l) = 20 µm.
-

Figure 29.
Alternaria phytolaccae (MFLUCC 21-0314, holotype). (a) Colonies on dead standing stem of Phytolacca sp. (b)–(e) Conidiophores bearing conidiogenous cells, with 2–3 conidiogenous loci on side of cell. (f), (g) Immature conidia. (h)–(k) Conidia. Scale bars: (a) = 150 µm, (b)–(k) = 20 µm.
-
Alternaria sections Conidiogenesis structures Ecology and economy References Sect. Alternantherae Conidiophores Short to moderately long, with slightly enlarged conidiogenous tip. Species in this section are reported as plant pathogens that mainly cause leaf spots. [11,13,15,55] Conidia Large, ellipsoidal to ovoid, or subcylindrical, rarely narrow ellipsoidal, solitary or rarely paired, disto- and euseptate, transversely septate with no or 1–2 longitudinal or oblique septa, slightly constricted near some septa, with a long apical narrow beak, conidial beak unbranched, septate or aseptate, long filiform, sometimes swollen at the end, internal compartmentation occurs, with cell bright at end, with hexagonal, octagonal or rounded transverse sections lumina. Sect. Alternaria Conidiophores Short to long, straight or curved, simple or branched, with one or several apical conidiogenous loci. Species in this section are reported as plant pathogens on leaves, stems and fruits, and vegetables. Some species cause opportunistic infections of humans. Species in this section are also reported as resources of potential toxins and secondary metabolites. [11,12,15,100] Conidia Obclavate to long ellipsoid, small or moderate in size, septate, slightly constricted near some septa, with few longitudinal septa, in moderately long to long, simple or branched chains, form tapered beak or secondary conidiophore with one or a few conidiogenous loci. Sect. Brassicicola Conidiophores Short to moderately long, simple or branched, with one or several apical conidiogenous loci. Species in this section mainly cause black spot disease on a wide range of hosts, particularly on Brassica spp. such as cabbage, Chinese cabbage, cauliflower, oilseeds, broccoli and canola. Species in this section are also reported as sources of antibiotic masses. [11,14,101−103] Conidia Ellipsoid, ovoid or somewhat obclavate, small or moderate in size, septate, slightly or strongly constricted at most of the transverse septa, with or without longitudinal septa, in moderately long to long, simple or branched chains, with dark septa and cell walls. Apically or laterally form secondary conidiophores with one or a few conidiogenous loci. Sometimes produced chlamydospores. Sect. Chalastospora Conidiophores Short to long, simple or branched, with one or several conidiogenous loci. Species in this section are primarily reported as saprobes and causal agents of human diseases. [11,30,38] Conidia Pale to medium brown, narrowly ellipsoidal to ellipsoidal or ovoid, beakless, with no or multiple transverse eusepta and rarely longitudinal septa, solitary or in chains. Apically or laterally form secondary conidiophores with one or a few conidiogenous loci. Sect. Cheiranthus Conidiophores Short to moderately long, simple or branched, with one or several conidiogenous loci. Species in this section are primarily saprobes and pathogens on various plant hosts. [11,38,55] Conidia Ovoid, broadly ellipsoid with transverse and longitudinal septa, slightly or strongly constricted at the septa, in short to long, simple or branched chains. Sect. Crivellia Conidiophores Straight or curved, simple or branched, with geniculate, sympodial proliferations. Species in this section are mainly known as pathogens on opium poppy (Papaver somniferum L.), the sexual morph of which links with genus Crivella. [11,58] Conidia Cylindrical, straight to curved to inequilateral, with transverse septa, rarely constricted at septa, single or in short, simple or branched chains. Apically or laterally form secondary conidiophores. Sometimes produced microsclerotia or chlamydospores. Sect. Dianthicola Conidiophores Simple or branched, with or without apical geniculate proliferations. Species in this section mainly cause leaf spot and blight on economic vegetation hosts such as carnation (Dianthus sp.) and sesame (Sesamum indicum L.). [11,104,105] Conidia Narrowly ovoid or narrowly ellipsoid with transverse and few longitudinal septa, slightly constricted at the septa, with a long (filamentous) beak or apical secondary conidiophore, solitary or in short chains. Sect. Embellisia Conidiophores Simple, septate, straight or with geniculate sympodial proliferation. Species in this section are reported as pathogens on vegetable crops such as tomato and garlic. [11,106,107] Conidia Solitary, ovoid to subcylindrical, straight to inequilateral, with transverse septa; septa can be thick, dark and rigid in contrast to the external wall. Sometimes sporulated chlamydospores. Sect. Embellisioides Conidiophores Simple, septate conidiophores, straight or with multiple, geniculate, sympodial proliferations. Species in this section are mainly reported as saprobes in soil and pathogen on plant hosts. [9,108,109] Conidia Solitary or in short chains, obovoid to ellipsoid, with transverse and longitudinal septa, transverse septa can be thick, dark and rigid in contrast to the external wall. Apical or lateral, short secondary conidiophores may occur. Sometimes produced sexual morph and chlamydospores. Sect. Eureka Conidiophores Simple, septate conidiophores, straight or with geniculate, sympodial proliferations. Species in this section are reported as pathogens and endophytes that are active in the biotransformation of some secondary metabolites. [11,110,111] Conidia Solitary or in short chains, narrowly ellipsoidal to cylindrical, with transverse and longitudinal septa, slightly constricted at the septa, with a blunt rounded apex. Sometimes form apical or lateral, short secondary conidiophores and sporulated sexual morph and chlamydospores. Sect. Euphorbiicola Conidiophores Short to long, broad, apical and sometimes lateral, secondary conidiophores. Species in this section served as pathogens on economic plants such as Euphorbiicola sp. and Citrus sp. and also produced secondary metabolites. [11,112] Conidia Medium to large-sized, in short to moderately long chains, ovoid, obclavate, disto- and euseptate, with multiple transverse and some longitudinal septa, slightly constricted near some transverse septa, with no or a simple long beak in the terminal conidia. Sect. Gypsophilae Conidiophores Simple, or occasionally branched, with one or a few conidiogenous loci. Species in this section occur on the host family Caryophyllaceae. [11,14,38] Conidia Solitary or in short chains, ellipsoid to long ovoid, with multiple transverse and longitudinal septa, conspicuously constricted near some transverse septa. Apically form secondary conidiophores with one or two conidiogenous loci or laterally with a single conidiogenous locus. Sect. Helianthiinficientes Conidiophores Simple, or branched, with one or a few conidiogenous loci. Species in this section is well-known as a pathogen on sunflower and cosmos, and also associated with some other species in Asteraceae (i.e., Arctium sp. and Sonchus sp.). [6] Conidia Solitary or in short chains, large, narrowly or broadly ovoid, or ellipsoidal, with several transverse and longitudinal septa, constricted near septa, sometimes non-beaked. Apically form secondary conidiophores, or a few lateral secondary conidiophores, or short to very long filiform beak. Sect. Infectoriae Conidiophores Short to long, simple or branched, with one or several conidiogenous loci. Species in this section are known as saprobes as well as plant and human pathogens. [11,14,38,
70]Conidia Moderately long to long, branched chains, small or moderate sized, obclavate to long ellipsoidal, septate, slightly constricted near some septa, with few longitudinal septa. Apically or laterally formed long geniculate, multi-locus secondary conidiophores, with meristematic growth. Sect. Japonicae Conidiophores Short to long, simple or occasionally branched, with a single conidiogenous locus. Species in this section particularly occur on hosts in Brassicaceae. [11,14] Conidia Short to long ovoid with transverse and longitudinal septa, conspicuously constricted at most of the transverse septa, in short chains. Apically formed secondary conidiophores with single conidiogenous locus. Sect. Nimbya Conidiophores Simple, short to form moderately long, sometimes one to a few short to long, geniculate, sympodial metastasis. Species in this section are known as saprobes and plant pathogens. Species in this section produce phytotoxins [11,55,85,
113,114]Conidia Solitary or in short chains, narrowly elongate-obclavate, gradually tapering apically, with transverse disto- and eusepta, sometimes slightly constricted near eusepta. Sect. Omanenses Conidiophores Long, simple, with multiple geniculate, sympodial metastasis or short conidiogenous loci normally with a terminal cluster of three conidia. Species in this section consist of a core taxon A. omanensis which is saprobic on dead woods. [7] Conidia Solitary, obovoid and sphaeroid, non-beaked, with transverse and longitudinal septa. Sect. Panax Conidiophores Simple or branched, short to moderately long, with one or a few conidiogenous loci. Species in this section are known as pathogens causing blight on economic plants such as ginseng and American ginseng (Araliaceae). [11,14,115] Conidia Solitary, simple or branched, in short chains, obclavate to ovoid, with multiple transverse and longitudinal septa, conspicuously constricted near several transverse septa, apically formed secondary conidiophores with one or several conidiogenous loci, multiple lateral secondary conidiophores with a single conidiogenous locus. Sect. Phragmosporae Conidiophores Simple, short to moderately long, with one or multiple geniculate, sympodial metastasis. Species in this section are mainly known as saprobes from soil and marine environments. [11] Conidia Solitary or in simple short chains, broadly ovoid to long ovoid, ellipsoidal, curved, or limaciform, with multiple transverse and few to multiple longitudinal septa, some septa darkened, slightly to conspicuously constricted near several transverse septa, apically formed secondary conidiophores with one or several conidiogenous loci. Sect. Porri Conidiophores Short to long, simple, with one or several conidiogenous loci. Species in this section consist of some important phytopathogens and produce phytotoxins. [11,14,22,116,117] Conidia Solitary or in short to moderately long chains, with a simple or branched, long to filamentous beak, medium or large size, broadly ovoid, obclavate, ellipsoid, subcylindrical or obovoid, disto- and eusepta, with multiple transverse and longitudinal septa, slightly constricted near some transverse septa, apically or laterally formed secondary conidiophores. Sect. Pseudoalternaria Conidiophores Simple or branched, septate, smooth, medium brown, simple with a single apical pore, with short to long, simple to multi-geniculate secondary conidiophores with one to many conidiogenous loci. Species in this section are known as pathogens on plant hosts. [15] Conidia Mostly catenulate, ellipsoid to obclavate, medium brown to golden brown, with several transverse and longitudinal septa, smooth, secondary conidiophore may occur as a false beak. Sect. Pseudoulocladium Conidiophores Simple or branched, with short, geniculate, sympodial metastasis. Species in this section are reported as phytopathogens for human infection. [11] Conidia Obovoid, non-beaked with a narrow base, in simple or mostly branched chains, apically formed secondary conidiophores with multiple conidiogenous loci and laterally secondary conidiophores may occur with a single conidiogenous locus. Sect. Radicina Conidiophores Straight, simple or branched, short or long, with multiple, short geniculate, sympodial proliferations, with one to a few conidiogenous loci at the apex. Species in this section mainly occur on hosts in family Apiaceae. [11] Conidia Solitary or in short chains, moderate in size, broadly ovoid to narrowly ellipsoidal, beakless, with several transverse and longitudinal septa, apically formed solitary, short, secondary conidiophores. Sect. Soda Conidiophores Simple or occasionally branched, short to moderately long, with one conidiogenous locus. Species in this section are isolated from soda lake environments (Western Siberia, Russia). [56] Conidia Solitary or in short to long, simple or branched chains, moderate to very large in size, narrowly ellipsoid to elongate-ovoid or somewhat obclavate, septate, with transverse and longitudinal septa, conspicuously constricted at most of the transverse septa, produced microsclerotia or chlamydospores, apical or lateral short secondary conidiophores with a single conidiogenous locus may occur, and conidiogenous tip can be enlarged. Sect. Sonchi Conidiophores Simple or branched, with short, geniculate, with one or several conidiogenous loci. Species in this section mainly occur on a wide range of hosts within Asteraceae (Compositae). [14] Conidia Single or in short chains, medium to large size, subcylindrical, broadly ovoid, broadly ellipsoid or obclavate, with multiple transverse and few longitudinal septa, slightly constricted at the septa. Sect. Teretispora Conidiophores Simple, sometimes extending at the apex with one or two, geniculate, sympodial proliferations. Species in this section consist of a core species, Alternaria leucanthemi, which is a phytopathogen causing plant blight disease. [11,38] Conidia Single, long cylindrical, lacking a beak portion, with many transverse and a few longitudinal septa, constricted at most of the transverse septa, secondary conidiophores with single conidium from the base of primary conidium and rarely formed apically. Sect. Ulocladioides Conidiophores Short, geniculate, sympodial proliferations. Species in this section are mainly known as phytopathogens causing leaf spot disease and can be saprobes on a variety of host substrates as well as a causal agent of keratitis. [11,15] Conidia Obovoid, non-beaked with a narrow base, single or in chains, with apical secondary conidiophores. Sect. Ulocladium Conidiophores Simple, with one or two short, geniculate, sympodial proliferations. Species in this section are mainly isolated from plant litter and rarely from marine environments. Potential bioactivities were also reported. [11,118] Conidia Single, obovoid, non-beaked, with a narrow base. Sect. Undifilum Conidiophores Simple, septate, straight, or with geniculate sympodial proliferation. Species in this section mainly occur on hosts in family Fabaceae. [11] Conidia Ovate to obclavate to long ellipsoid, straight to inequilateral, single, transverse septa, septa can be thick, dark and rigid, and form unique germ tubes, which are wavy or undulate until branching. Table 1.
Synopsis of Alternaria sections based on the asexual morphs.
-
Gene loci Primers Sequence 5’–3’ References Internal transcribed spacer region (ITS, including the 5.8S gene) ITS5 GGA AGT AAA AGT CGT AAC AAG G [139] ITS4 TCC TCC GCT TAT TGA TAT GC 28S large subunit rDNA (LSU) LR0R GTA CCC GCT GAA CTT AAG C [140] LR5 ATC CTG AGG GAA ACT TC 18S small subunit rDNA (SSU) NS1 GTA GTC ATA TGC TTG TCT C [139] NS4 CTT CCG TCA ATT CCT TTA AG Alternaria major allergen (Alt-a1) Alt-for ATG CAG TTC ACC ACC ATC GC [49] Alt-rev ACG AGG GTG AYG TAG GCG TC Glyceraldehyde 3-phosphate Dehydrogenase (GAPDH) GDP-1 CAA CGG CTT CGG TCG CAT TG [141] GDP-2 GCC AAG CAG TTG GTT GTG C Plasma membrane ATPase (ATPase) ATPDF1 ATC GTC TCC ATG ACC GAG TTC G [14] ATPDR1 TCC GAT GGA GTT CAT GAT AGC C The second largest subunit of RNA polymerase II (RPB2) fRPB2-5f GAY GAY MGW GAT CAY TTY GG [142] fRPB2-7cR CCC ATR GCT TGY TTR CCC AT Translation elongation factor 1-α (TEF1-α) EF1-983F GCY CCY GGH CAY CGT GAY TTY AT [143] EF1-2218R ATG ACA CCR ACR GCR ACR GTY TG EF1-728F CATCGAGAAGTTCGAGAAGG [144] EF1-986R TACTTGAAGGAACCCTTACC Table 2.
Gene loci and primers used in this study.
-
Phylogenetic analyses Nucleotide substitution models ITS LSU SSU GAPDH RPB2 TEF1-α Alt-a1 ATPase A1: Alternaria sections GTR+I+G GTR+G TrN+I+G SYM+I+G GTR+I+G TIM1+I+G GTR+I+G n/a A2: A. alternata GTR+I+G GTR+I+G GTR+I+G GTR+I+G TIM2 +G GTR+I+G GTR+I+G n/a A3: sect. Infectoriae GTR+I+G n/a n/a GTR+I+G n/a n/a n/a SYM+G A4: sect. Porri SYM+I+G n/a n/a TIM2+I+G GTR+I+G GTR+G GTR+I+G n/a A5: sect. Radicina GTR+I+G n/a n/a GTR+I+G TIM2+G GTR+I+G n/a n/a Table 3.
The best nucleotide substitution model for each locus based on the Akaike Information Criterion (AIC) generated by MrModeltest v. 2.3.[156].
-
Order Family Genus Sections Divergence times (crown age) Divergence times (stem age) Pleosporales 233 (168–301) Mya 252 (184–326) Mya Pleosporaceae 110 (79–148) Mya 120 (84–159) Mya Alternaria 53 (36–71) Mya 62 (42–85) Mya Alternaria sect. Alternantherae 0.4 (0–1.5) Mya 14 (6.7–21) Mya Alternaria sect. Alternaria 5 (1.7–10) Mya 14 (6.7–21) Mya Alternaria sect. Brassicicola 2.3 (0.5–5.5) Mya 33 (22–45) Mya Alternaria sect. Chalastospora 16 (8.8–26) Mya 26 (16–38) Mya Alternaria sect. Cheiranthus 11 (4.23–20) Mya 26 (16–38) Mya Alternaria sect. Crivellia 7.6 (1.5–19) Mya 53 (36–71) Mya Alternaria sect. Dianthicola 11 (5.4–18) Mya 17 (10–27) Mya Alternaria sect. Embellisia 7.4 (2.5–15) Mya 28 (14–43) Mya Alternaria sect. Embellisioides 11 (5–19) Mya 24 (14–36) Mya Alternaria sect. Eureka 14 (5.6–24) Mya 28 (18–44) Mya Alternaria sect. Euphorbiicola - 11 (5.6–17) Mya Alternaria sect. Gypsophilae 16 (7.6–26) Mya 27 (18–37) Mya Alternaria sect. Helianthiinficientes 0.11 (0–0.3) Mya 24 (13–36) Mya Alternaria sect. Infectoriae 9.5 (4–17) Mya 26 (16–38) Mya Alternaria sect. Japonicae – 31 (14–47) Mya Alternaria sect. Nimbya 24 (11–39) Mya 36 (28–51) Mya Alternaria sect. Omanenses – 30 (14–47) Mya Alternaria sect. Panax 14 (6.8–23) Mya 22 (12–33) Mya Alternaria sect. Phragmosporae 28 (13–44) Mya 42 (28–58) Mya Alternaria sect. Porri 6.7 (3–11) Mya 11 (5.6–17) Mya Alternaria sect. Pseudoalternata – 28 (14–43) Mya* Alternaria sect. Pseudoulocladium 2.1 (0.4–5.8) Mya 17 (9.5–27) Mya Alternaria sect. Radicina 9.3 (3.6–18) Mya 29 (19–40) Mya Alternaria sect. Soda 3 (0.5–8.4) Mya 32 (26–54) Mya Alternaria sect. Sonchi 6.8 (2–14) Mya 24 (13–36) Mya Alternaria sect. Teretispora 0.2 (0–1.03) Mya 27 (17–40) Mya Alternaria sect. Ulocladioides 8.1 (3–17) Mya 22 (13–32) Mya Alternaria sect. Ulocladium 0.9 (0.1–2.5) Mya 44 (32–60) Mya Alternaria sect. Undifilum – 45 (32–62) Mya Table 4.
Divergence times of Alternaria sections indicated in MCC tree. The age value with '*' indicates recent results lacking key coding gene strains.
-
Culture collection Original
codeHerbarium no. Origin Host and habitat Collection date Collector KUNCC 22-10823 IT2053 HKAS 124866
MFLU 15-2585Italy, Province of Forlì-Cesena, Premilcuore Dead stem of Sonchus sp. (Asteraceae) 18 August 2014 E. Camporesi KUNCC 22-10824 IT2063 HKAS 124867 Italy, Province of Forlì-Cesena, Verghereto, Montecoronaro Dead hanging stem of Centaurea sp. (Asteraceae) 20 August 2014 E. Camporesi KUNCC 22-10825 IT2064 HKAS 124868 Italy, Province of Forlì-Cesena, Fiumicello di Premilcuore Dead hanging stem of Helleborus sp. (Ranunculaceae). 28 August 2014 E. Camporesi KUNCC 22-10826 IT2087 HKAS 124869 Italy, Province of Forlì-Cesena, Quattro di Forlì Dead hanging stem of Agropyron sp. (Asteraceae) 1 September 2014 E. Camporesi KUNCC 22-10827 IT2090 HKAS 124870 Italy, Province of Forlì-Cesena, Predappio, Rocca delle Caminate Dead leaf petiole of Robinia sp. (Fabaceae) 4 September 2014 E. Camporesi KUNCC 22-10828 IT2103 HKAS 124871 Italy, Province of Forlì-Cesena, Meldola Dead hanging stem of Echinochloa
sp. (Poaceae)8 September 2014 E. Camporesi KUNCC 22-10829 IT2114 HKAS 124872 Italy, Province of Forlì-Cesena, Tessello Dead hanging stem of Cephalaria sp. (Dipsacaseae) 16 September 2014 E. Camporesi KUNCC 22-10830 IT2115 HKAS 124873 Italy, Province of Forlì-Cesena, Civitella di Romagna Dead hanging stem of Aster sp. (Asteraceae) 19 September 2014 E. Camporesi KUNCC 22-10831 IT2125 HKAS 124874 Italy, Province of Arezzo, Stia, Montemezzano Dead hanging stem of Euphorbia sp. (Euphorbiaceae) 22 September 2014 E. Camporesi KUNCC 22-10832 IT2143 HKAS 124875 Italy, Province of Forlì-Cesena, Galeata, San Zeno Dead hanging leaf petiole of
Ailanthus sp. (Simaroubaceae)30 September 2014 E. Camporesi KUNCC 22-10833 IT2144 HKAS 124876 Italy, Province of Forlì-Cesena,
Cabelli di Santa SofiaDead hanging stem of Agrostis sp. (Poaceae) 2 October 2014 E. Camporesi KUNCC 22-10834 IT2145 HKAS 124877 Italy, Province of Forlì-Cesena,
Monte MirabelloDead hanging leaf of Arundo sp. (Poaceae) 3 October 2014 E. Camporesi KUNCC 22-10835 IT2162 HKAS 124878 Italy, Province of Forlì-Cesena,
Santa SofiaDead hanging stem of Hedysarum coronarium L. (Papilionaceae) 7 October 2014 E. Camporesi KUNCC 22-10836 IT2166 HKAS 124879 Italy, Province of Forlì-Cesena, Meldola, Piandispino Dead hanging stem of Dipsacus sp. (Caprifoliaceae) 7 October 2014 E. Camporesi KUNCC 22-10837 IT2277 HKAS 124880 Italy, Province of Ravenna, Lido di Dante Dead hanging stem of Kali tragus (L.) Scop. (Amaranthaceae) 2 December 2014 E. Camporesi KUNCC 22-10838 IT2347 HKAS 124881 Italy, Province of Forlì-Cesena,
Collina di ForlìSeveral samaras of Fraxinus oxycarpa Willd. (Oleaceae) 21 January 2015 E. Camporesi KUNCC 22-10839 IT2427 MFLU 15-1823 Italy, Province of Forlì-Cesena, Forlì, Via Nenni Dead hanging stem of Wisteria sp. (Caprifoliaceae) 30 March 2015 E. Camporesi KUNCC 22-10840 IT2454 HKAS 124883 Italy, Province of Forlì-Cesena, Predappio, Rocca delle Caminate Dead stem of Urtica sp. (Urticaceae) 21 April 2015 E. Camporesi KUNCC 22-10841 IT2900 MFLU 16-1116 Italy, Province of Forlì-Cesena, Santa Sofia, Camposonaldo Dead needles of Pinus nigra
J.F. Arnold (Pinaceae)23 March 2016 E. Camporesi KUNCC 22-10842 IT3048 MFLU 16-2277 Italy, Province of Forlì-Cesena, Fiumicello di Premilcuore Dead hanging stem of Acer opalus Mill. (Sapindaceae) 27 July 2016 E. Camporesi KUNCC 22-10843 IT3160 MFLU 16-2883 Italy, Province of Forlì-Cesena, Forlì, Ravaldino in Monte Dead stem of Silybum marianum (L.) Gaertn. (Asteraceae) 15 November 2016 E. Camporesi KUNCC 22-10844 IT3168 MFLU 16-2904 Italy, Province of Forlì-Cesena, Forlì, Parco Urbano Dead hanging fruits of Ostrya carpinifolia Scop. (Betulaceae) 19 November 2016 E. Camporesi KUNCC 22-10845 IT3419 HKAS 124888 Italy, Province of Arezzo, Poppi, Quota Dead hanging stem of Calamintha nepeta (Lamiaceae) 25 July 2017 E. Camporesi KUNCC 22-10846 IT3426 HKAS 124889 Italy, Province of Arezzo, near Croce di Pratomagno Dead hanging stem of Malva alcea L. (Malavaceae) 1 August 2017 E. Camporesi KUNCC 22-10847 IT3439 HKAS 124890 Italy, Province of Arezzo, Stia, Montemezzano Dead hanging stem of Reseda luteola L. (Resedaceae) 13 August 2017 E. Camporesi KUNCC 22-10848 IT3442 HKAS 124891 Italy, Province of Arezzo, Montemignaio Dead hanging stem of Valeriana sp. (Caprifoliaceae) 12 August 2017 E. Camporesi KUNCC 22-10849 IT3449 HKAS 124892 Italy, Province of Forlì-Cesena, Bagno di Romagna, Riofreddo Dead hanging stem of Tanacetum
sp. (Asteraceae)22 August 2017 E. Camporesi KUNCC 22-1050 IT3504 MFLU 17-1778 Italy, Province of Forlì-Cesena, Bagno di Romagna, Acquapartita Dead hanging stem of Orobanche
sp. (Orobanchaceae)25 September 2017 E. Camporesi KUNCC 22-10851 IT3556 HKAS 124894 Italy, Province of Forlì-Cesena, Santa Sofia Dead leaf of Sorbus aria Crantz (Rosaceae) 15 November 2017 E. Camporesi KUNCC 22-10852 IT3598 HKAS 124895 Italy, Province of Ravenna, Faenza, Santa Lucia Dead leaves of Quercus pubescens Willd. (Fagaceae) 13 December 2017 E. Camporesi KUNCC 22-10853 IT3651 HKAS 124896 Italy, Province of Forlì-Cesena, Meldola Dead hanging branch of Sambucus nigra L. (Adoxaceae) 31 December 2017 E. Camporesi KUNCC 22-10854 KIB-H2 HKAS 124897 China, Yunnan, Kunming Institute of Botany Dead fallen leave of bamboo (Poaceae) 26 December 2014 J.F. Li KUNCC 22-10855 HKM-1 HKAS 124898 China, Yunnan, Kunming, Xundian Dead stem of Raphanus sativus L. (Brassicaceae) 13 March 2015 J.F. Li KUNCC 22-10856 H-49 HKAS 124899 China, Yunnan, Baoshan Dead branch of Capsicum annuum L. (Solanaceae) 22 October 2015 J.F. Li KUNCC 22-10857 PB-12 HKAS 124900 China, Yunnan, Pingbian, Dawei Mountain Dead stem of Zea mays L. (Poaceae) 20 September 2017 J.F. Li KUNCC 22-10858 HXB-08 HKAS 124901 China, Yunnan, Xishuangbanna Dead fallen leaves of Dimocarpus longan Lour. (Sapindaceae) 8 June 2018, J.F. Li KUNCC 22-10859 HAM-02 HKAS 124902 China, Yunnan, Honghe, Amu Mountain Dead fallen leaves of grass (Poaceae) 15 June 2018 J.F. Li KUNCC 22-10860 HAM-03 HKAS 124903 China, Yunnan, Honghe, Amu Mountain Dead aerial stem of Capsicum
annuum (Solanaceae)15 June 2018 J.F. Li KUNCC 22-10861 HSH-01 HKAS 124904 Thailand, Chiang Rai, Muang, Singha Park Dead culms of bamboo (Poaceae) 23 February 2016 J.F. Li KUNCC 22-10862 HMRC-51 HKAS 124905 Thailand, Chiang Mai, Mae Taeng, Mushroom Research Center (M.R.C) Dead fallen leaves of unidentified plant 23 March 2016 J.F. Li KUNCC 22-10863 DMS-15 HKAS 124906 Thailand, Chiang Rai, Doi Mae Salong Dead leaves of grass (Poaceae) 24 May 2016 J.F. Li KUNCC 22-10864 HTWD-01 HKAS 124807 Thailand, Chiang Rai, Mae Fah Luang Dead leaves of palm (Arecaceae) 25 September 2016 J.F. Li KUNCC 22-10865 H-71 HKAS 124908 Thailand, Chiang Rai, Doi Chang Dead leaves of Coffea arabica L. (Rubiaceae) 25 July 2018 J.F. Li KUNCC 22-10866 HWP-01 HKAS 124909 Thailand, Chiang Rai, Wiang Pa Pao Dead stems of Bidens pilosa L. (Asteraceae) 16 October 2018 J.F. Li Table 5.
Additional collections of Alternaria alternata collected from Yunnan, China, Italy and Thailand in this study.
-
Species name Strain no. Nucleotide difference of gene sequences (no gaps) A. doliconidium MFLUCC 14-0020 ITS TEF1-α RBP2 GAPDH Alt-a1 A. alternata CBS 102595 8/520 bp (1.5%) 14/256 bp (4.7%) 30/875 bp (3.4%) 16/582 bp (2.7%) 12/476 bp (2.5%) A. alternata CBS 916.96 8/520 bp (1.5%) 14/256 bp (4.7%) 28/875 bp (3.2%) 18/582 bp (3.1%) 12/476 bp (2.5%) A. alternata CBS 175.52 10/520 bp (1.9%) 16/256 bp (6.3%) 30/875 bp (3.4%) 17/582 bp (2.9%) 12/476 bp (2.5%) A. alternata YL1 9/520 bp (1.7%) 14/256 bp (4.7%) 30/875 bp (3.4%) 16/582 bp (2.7%) 12/476 bp (2.5%) A. alternata YL2 9/520 bp (1.7%) 14/256 bp (4.7%) 30/875 bp (3.4%) 17/582 bp (2.9%) 12/476 bp (2.5%) A. doliconidium HKAS 100840 1/520 bp, (0.2%) 0% N/A N/A N/A A. doliconidium MFLUCC 17-0263 1/520 bp, (0.2%) 0% N/A N/A N/A A. italica MFLUCC 14-0231 6/520 bp, (1.2%) 11/256 bp (4.3%) 25/875 bp, (2.9%) N/A N/A A. italica KUMCC 17-0123 7/520 bp, (1.3%) N/A N/A N/A N/A Table 6.
Nucleotide base comparison of Alternaria doliconidium (MFLUCC 14-0020) with closely related taxa.
Figures
(29)
Tables
(6)