-

Figure 1.
The effects of ethylene treatment on fruit ripening in ‘Summer Black’ grape. (a) Ripening phenotype in response to ethylene treatment (on day 0), Bar = 0.5 cm. (b) Single berry weight. (c) Longitudinal diameter. (d) Transverse diameter. (e) Total soluble solids. (f) Titratable acid. (g) Anthocyanin content. (h) Chlorophyll content. (i) Ethylene production.
-
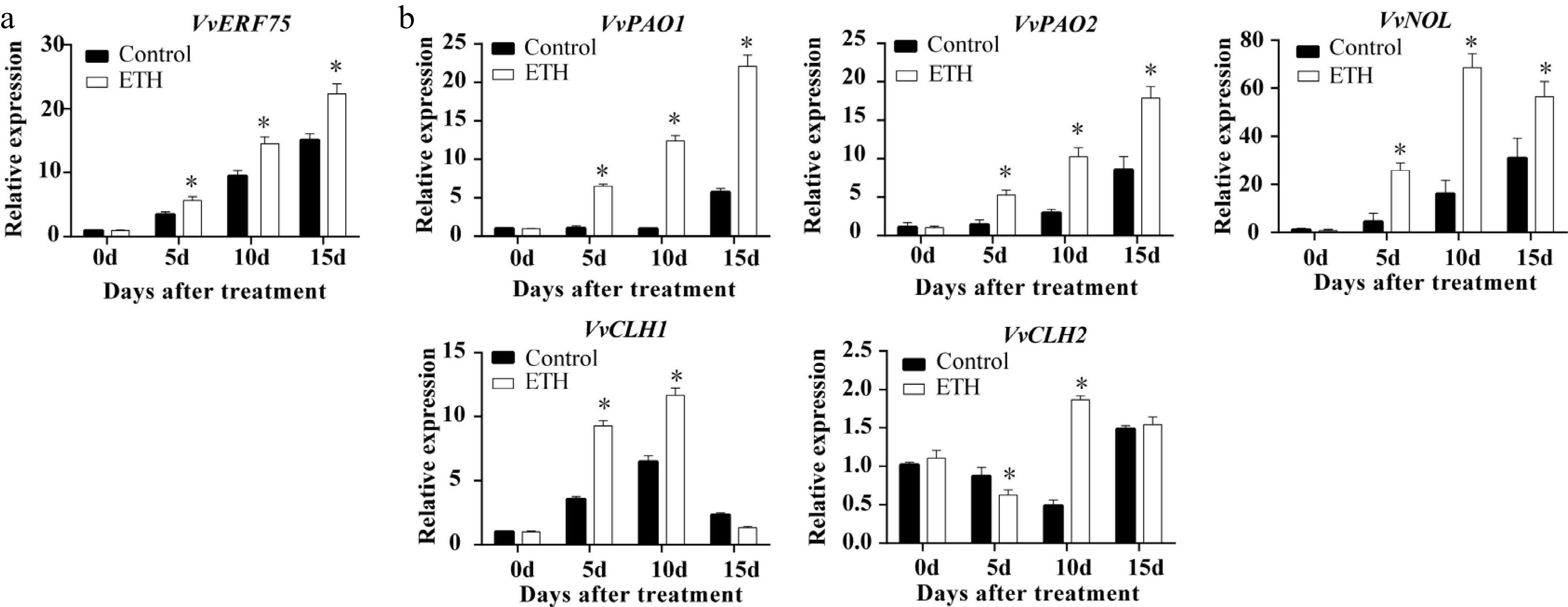
Figure 2.
Expression in ‘Summer Black’ grape of (a) VvERF75 and (b) chlorophyll degradation related genes in response to ethylene treatment.
-
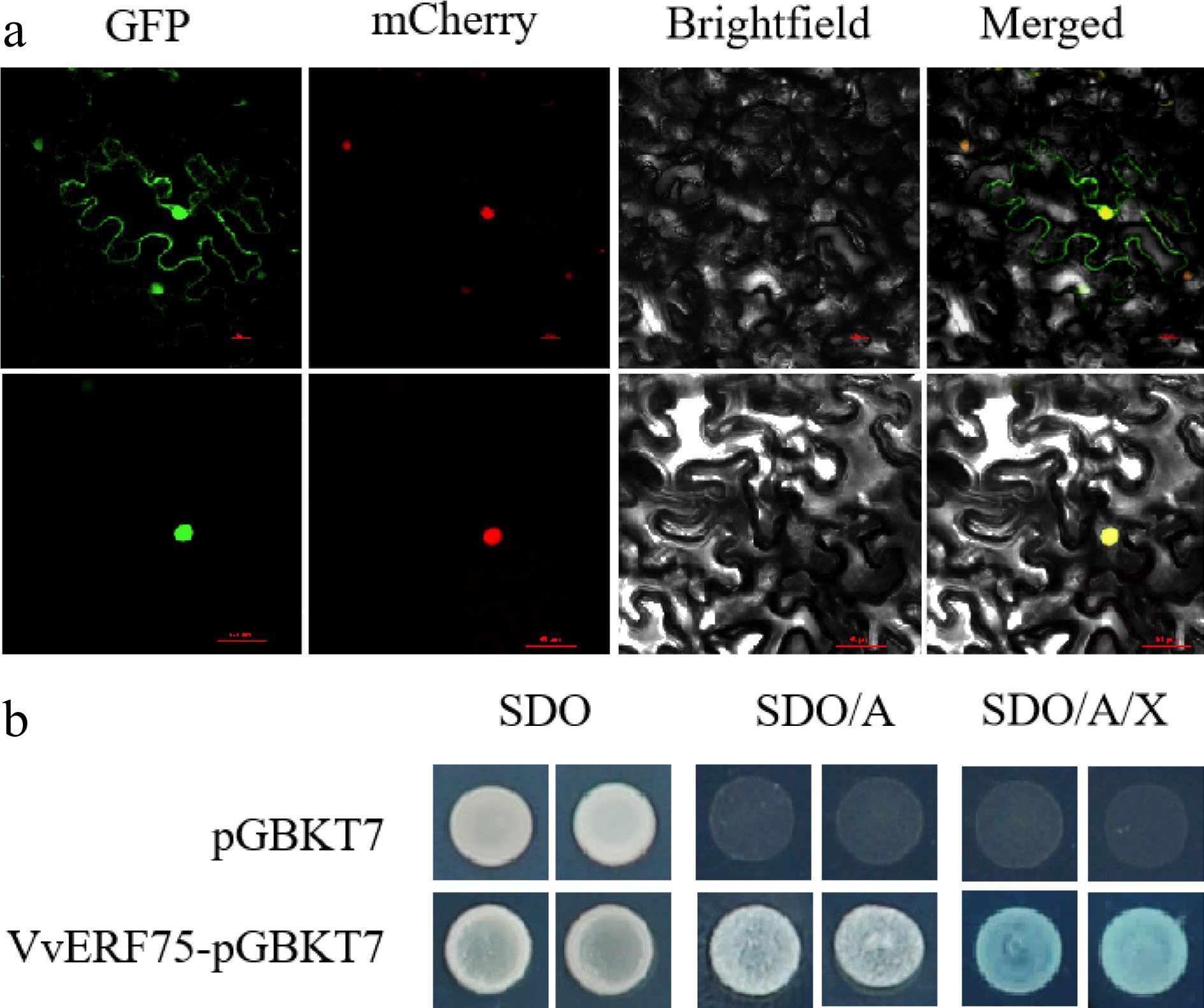
Figure 3.
Subcellular localization in tobacco and transcriptional activation in yeast of VvERF75. (a) Subcellular localization of VvERF75 in Nicotiana benthamiana leaves. VvERF75-GFP was transformed into Nicotiana benthamiana. VvERF75-GFP, GFP signal; Brightfield, white light; Merged, combined GFP and brightfield signals. NLS mCherry was used as a nuclear-localized marker. Bar = 50 μm. (b) VvERF75 trans-activation activity assay. Yeast cells were dotted on plates containing either SDO, synthetic dropout media; SDO/A, synthetic dropout medium supplemented with Aureobasidin A but lacking tryptophan; or SD/AbA/X, synthetic dropout medium supplemented with Aureobasidin A and X-a-Gal but lacking tryptophan. pGBKT7 vector was used as negative control.
-
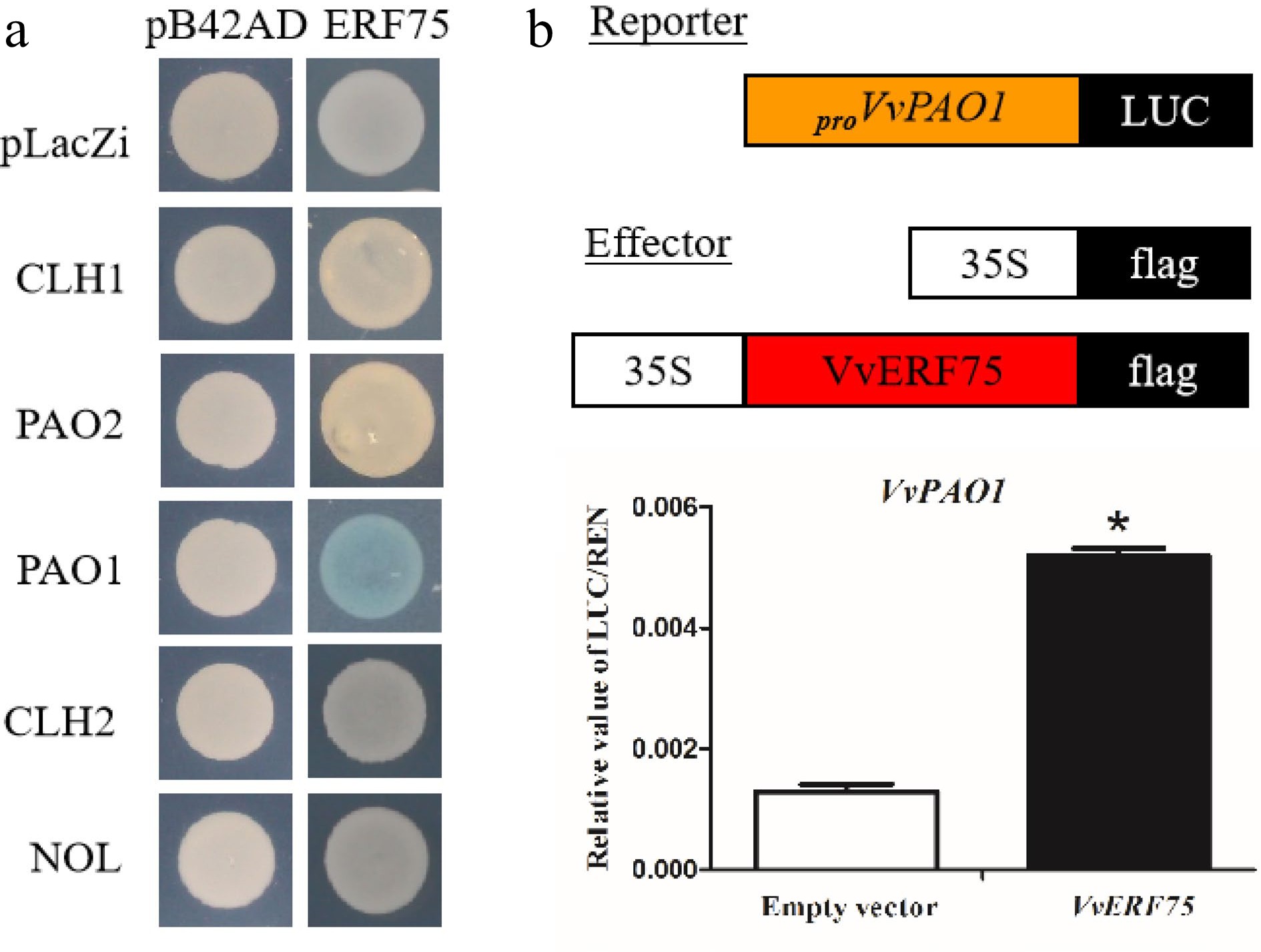
Figure 4.
Analysis of VvERF75 regulation of VvPAO1. (a) Yeast one-hybrid analysis of VvERF75 binding to the promoters of chlorophyll catabolic genes. (b) Schematics of the reporter and effector constructs used in dual-luciferase reporter assay. (c) Activation of the VvPAO1 gene by VvERF75 is indicated by the LUC/REN ratio. Empty vector: 35S:flag + proVvPAO:LUC; VvERF75: 35S:VvERF75:flag + proVvPAO:LUC, the empty effector was used as a control.
-
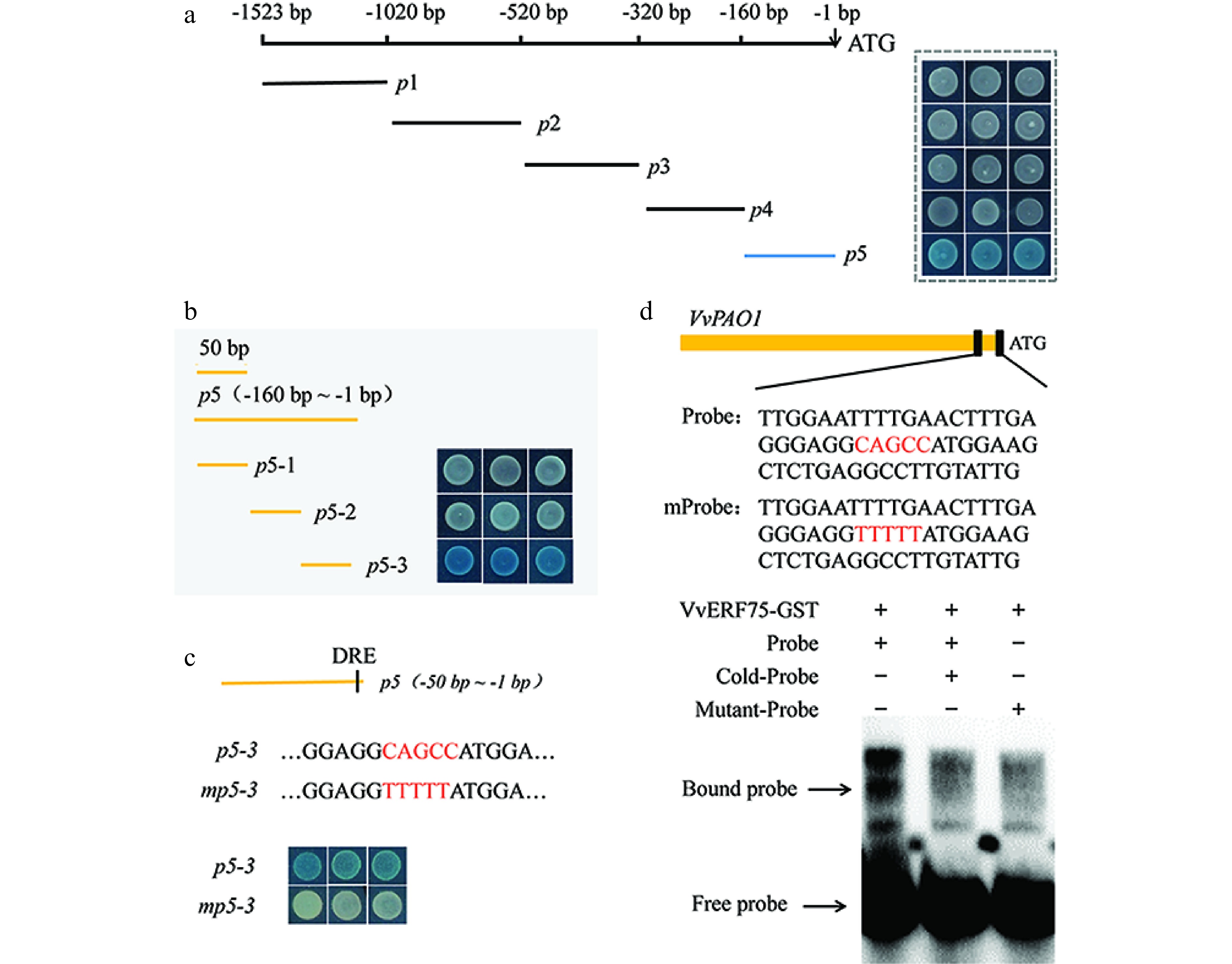
Figure 5.
Electromobility shift and yeast one-hybrid assays showing the binding of VvERF75 to the promoter of VvPAO1. (a), (b) Sequential deletion series and (c) mutation of VvPAO1 promoter for use in yeast one-hybrid assays with VvERF75. (d) Sequence of the probe derived from the promoter of VvPAO1 (p5-3) and VvPAO1m are shown, with the DRE motif and its mutant indicated in red. (e) EMSA of VvERF75 protein binding to the promoter sequence of the VvPAO1 gene. + and − represent the presence or absence of the respective composition in the binding experiment. The labeled 'free probe' and DNA protein complex 'bound probe' positions are indicated, showing competition with an unlabeled fragment for each promoter domain.
-
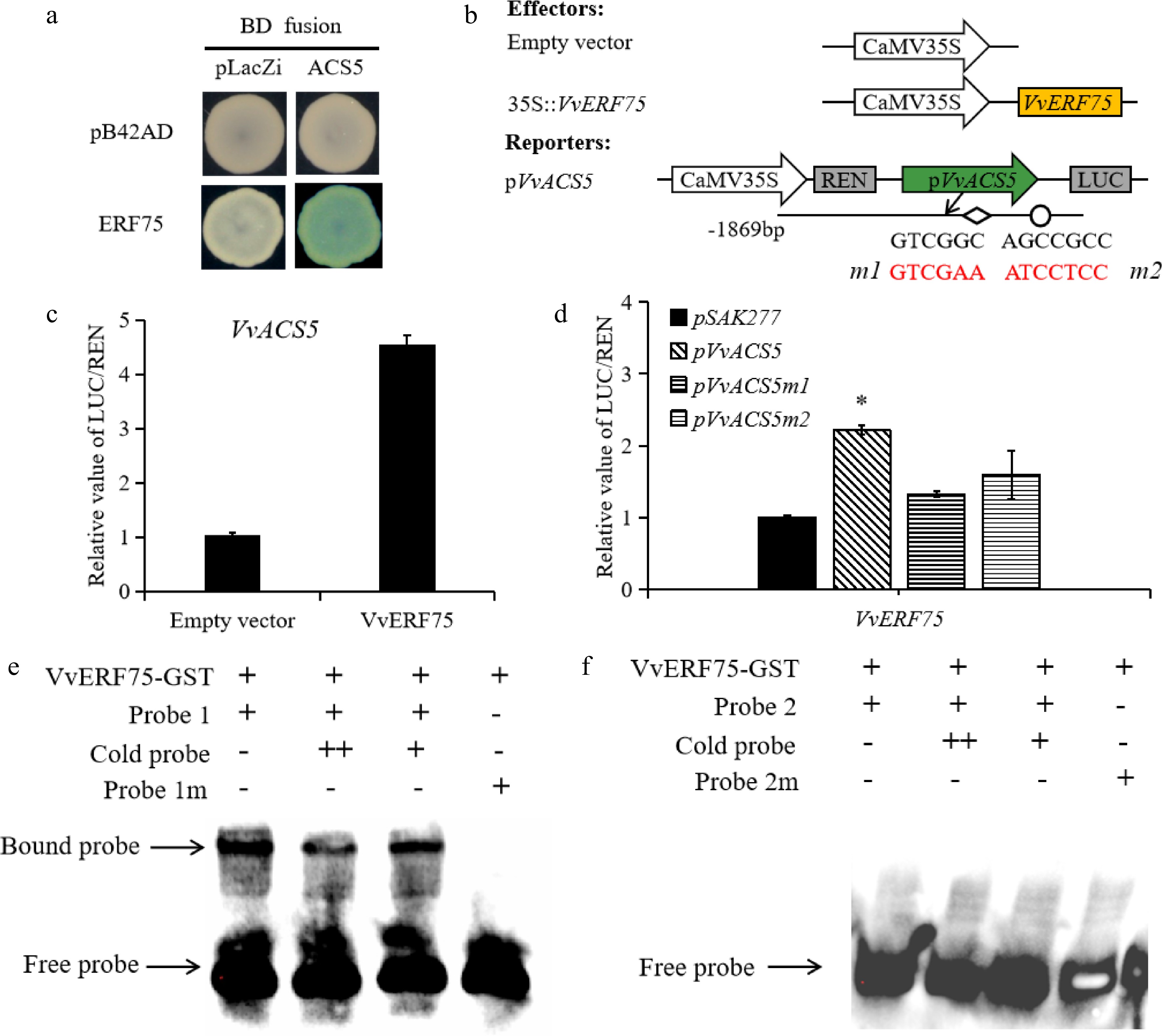
Figure 6.
Regulation of VvACS5 by VvERF75. (a) Yeast one-hybrid assay analysis of VvERF75 binding to VvACS5 promoter. (b) Schematics of the reporter and effector constructs used in the dual-luciferase reporter assay, the DRE motif and its mutant indicated in red. (c) Dual-luciferase assays of VvERF75 on promoter of VvACS5. (d) VvERF75 activation of the wild-type and mutant promoters of the VvACS5 genes. m1 carries a mutation of the DRE motif and m2 of the GCC box in the promoter of VvACS5. (e), (f) EMSA of VvERF75 protein binding to motifs 1 and 2 on promoter of the VvACS51 gene. + and − represent the presence or absence of the respective component in the binding experiment. The labeled 'free probe' and DNA protein complex 'bound probe' positions are indicated, showing competition with an unlabeled fragment for each promoter domain.
-
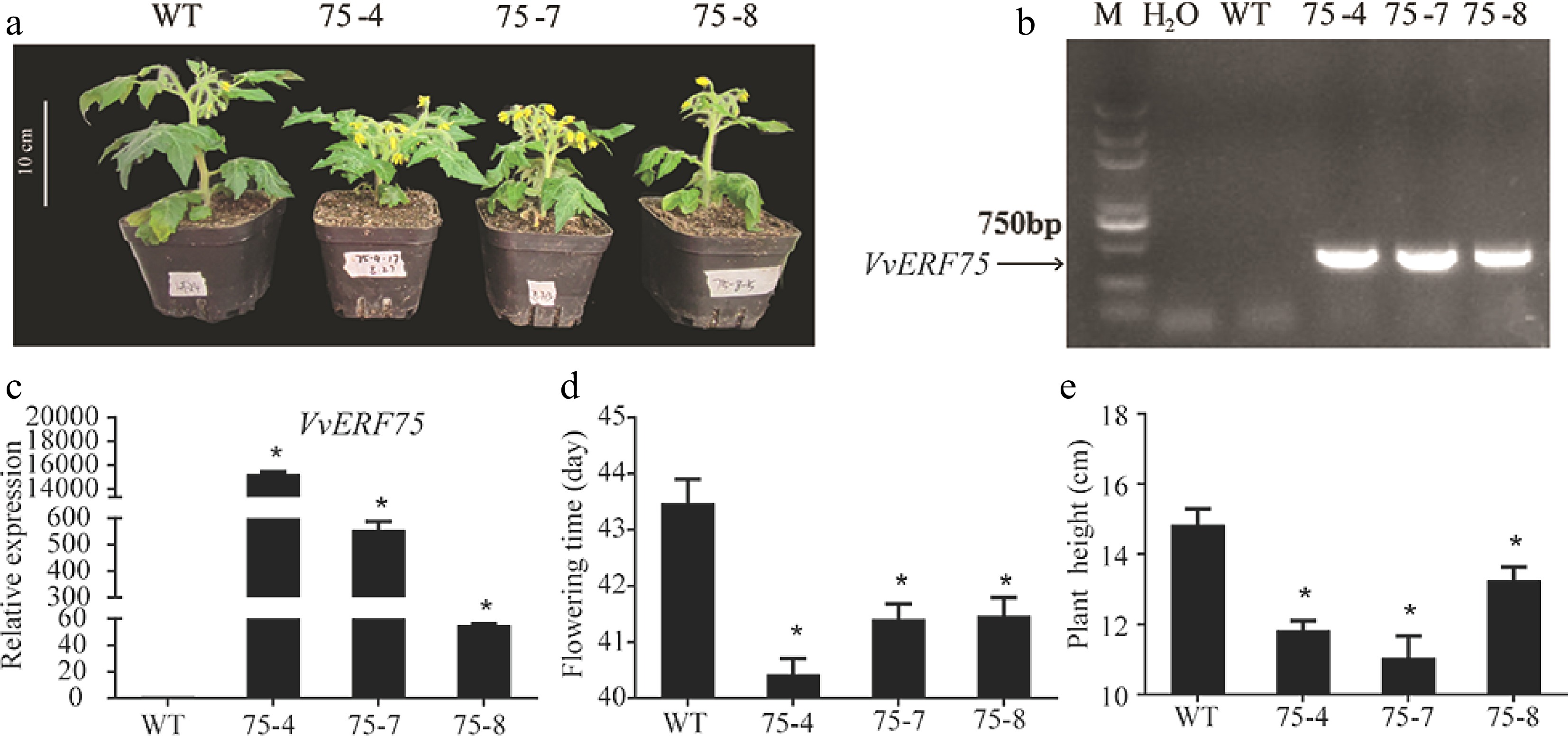
Figure 7.
Overexpression of VvERF75 in tomato changed flowering time and plant height. (a) Transgenic plants of VvERF75. Detection of transgenic VvERF75 plants by (b) PCR and (c) qRT-PCR. (d) Flowering time of transgenic VvERF75-OE plants compared to WT. (e) Height of transgenic VvERF75-OE plants compared to WT.
-
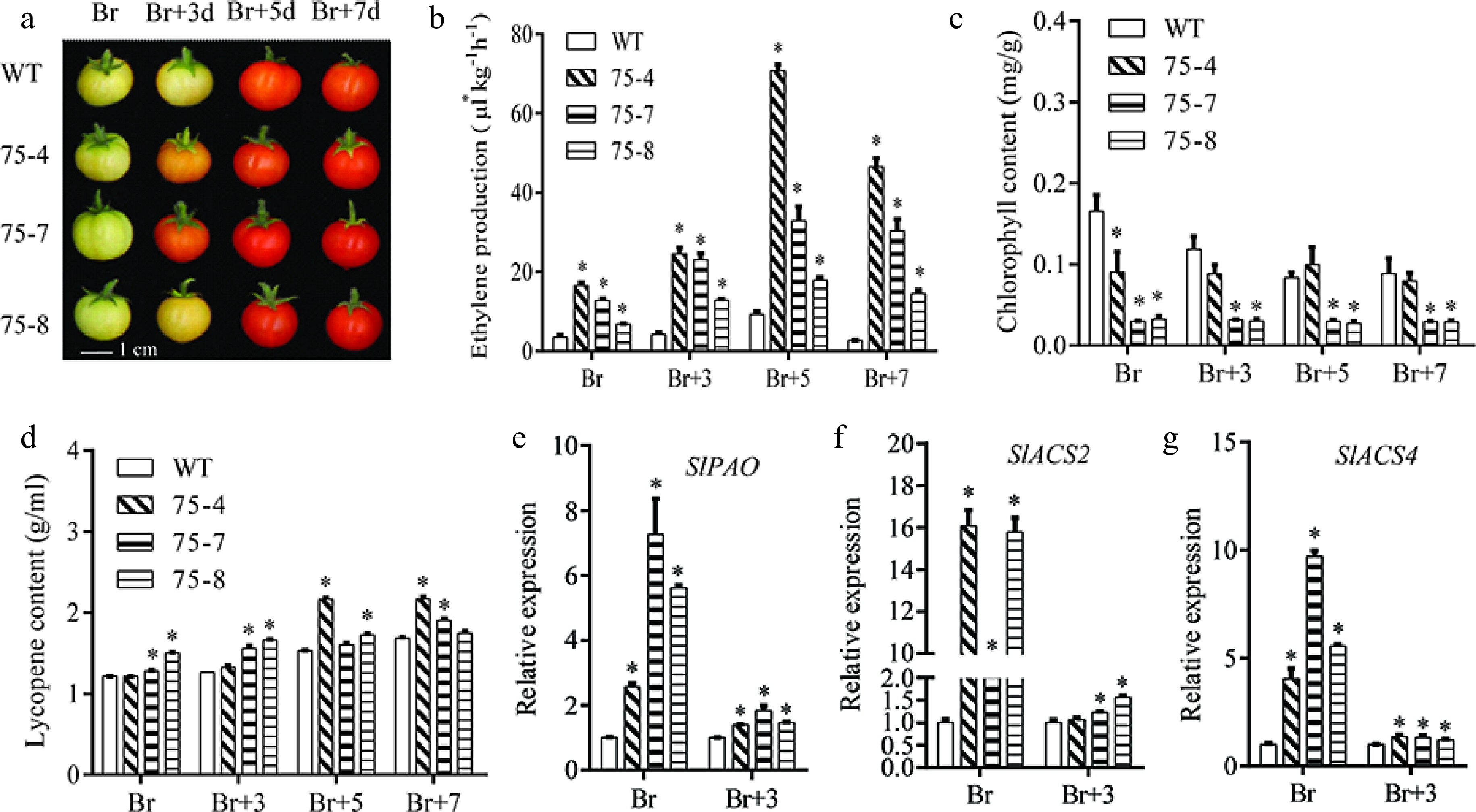
Figure 8.
VvERF75 enhanced ethylene production and chlorophyll degradation in tomato. (a) Color phenotype of VvERF75 overexpression lines. (b) Ethylene production, (c) chlorophyll content, (d) lycopene content in WT and VvERF75 overexpression lines at Br, Br+3, Br+5 and Br+7 stage. Expression of (e) SlPAO, (f) SlACS2 and (g) SlACS4 in WT and VvERF75-overexpression lines at Br and Br+3 stage.
Figures
(8)
Tables
(0)