-
Grape (Vitis vinifera L.) is an important economic fruit planted worldwide. Fruit ripening is crucial to the quality of a cluster of grapes, which affects both consumer selection and storability of the fruit. Fruit ripening is a complicated procedure that involves numerous physiological and biochemical reactions that result in color and textural changes, soluble sugar accumulation, and aroma formation in the fruit. Therefore, to improve fruit appearance, quality and storability, one must understand the regulatory mechanisms underlying fruit ripening and how these mechanisms are similar and different between fruit types.
Fleshy fruits are traditionally classified into climacteric and non-climacteric, according to whether there is a peak of respiration and ethylene release at the beginning of ripening. It has been widely studied how ethylene accelerates ripening of climacteric fruits[1−3]. Grape is a non-climacteric fruit, however, previous research implies that ethylene is required for grape ripening[4,5], that exogenous ethylene can affect coloration and ripening in non-climacteric fruit[6−8], and that ethylene treatment of grapes one week before veraison can enhance fruit color and anthocyanin content together with the expression of genes related to anthocyanin biosynthesis[8−10]. Less is known about the mechanism through which ethylene regulates grape berry ripening.
Grape berry ripening is firstly accompanied by changes in the color that result from chlorophyll degradation and anthocyanin accumulation[8−10]. Chl degradation has been widely investigated during leaf senescence and fruit ripening, resulting in the degreening of leaves or fruits[11−14], with the genes encoding the enzymes needed for the catalysis of Chl degradation identified, including NON-YELLOW COLOURING (NYC), PAO, chlorophyllase (CLH), pheophorbide hydrolase (PPH), Red chlorophyll catabolite reductase (RCCR) and STAYGREEN/STSYGREEN-LIKE (SGR/SGRL)[11,15−18]. Degradation of Chl during apple storage proved to be regulated by ethylene, which activates the expression of Chl catabolic genes (CCGs) that will result in accelerated Chl degradation[3,4,19].
Ethylene-induced Chl degradation is regulated transcriptionally[4,5,19] through activation of a downstream Ethylene Response Factor (ERF). ERFs belong to the AP2/ERF transcription factor superfamily[20] and regulate expression of CCGs during fruit ripening[4,5], leaf senescence, and iron deficiency[21,22], among other processes. Moreover, ERF transcription factors can modulate ethylene production by regulating expression of ACS and ACC oxidase (ACO), which encode key enzymes in ethylene biosynthesis[2,3,23,24]. Genome-wide identification of AP2/ERF genes in grape has been carried out, and 122 ERF TFs were identified, which can be classified into 12 subfamilies[25]. VvERF17 has been shown to control chlorophyll degradation in berry skin during fruit development[26], while VvERF95 was proven to regulate chlorophyll degradation during rachis degreening during postharvest storage. However, relatively few functional analyses of grape ERF genes have been reported, and it remains unknown if ERF transcription factors are involved in ethylene biosynthesis and Chl degradation during grape berry coloration.
In this study, VvERF75 is shown to have a higher expression level after ethylene treatment and a positive correlation with ethylene production, but a significantly negative correlation with chlorophyll content. We further studied the VvERF75-controlled regulation of CCGs and ethylene biosynthesis genes and demonstrated the functions of VvERF75 in Chl degradation and ethylene biosynthesis. The results of this study advance our understanding of the mechanisms through which ethylene influences grape ripening.
-
Vines of the cultivar ‘Summer Black' (Vitis vinifera × Vitis labrusca) were planted in Henan Agricultural University (China) and maintained under normal growth conditions. Ethylene (500 mg/L) was sprayed on berries one week before veraison, with berries sprayed with distilled water used as control. Nine grapevine clusters were sampled randomly at 0, 5, 10 and 15 days (d) after treatment. At each timepoint, 30 berries were randomly selected for measurement of longitudinal diameter, transverse diameter, single berry weight (mm), soluble solids content (%), and titratable acidity (%). Eighty grapevine berries were deseeded, immediately frozen in liquid nitrogen and stored at −80 ºC until assayed.
Gene isolation, phylogenetic tree construction, multiple sequence alignment and motif analysis of promoters
-
The full-length coding sequence (CDS) of VvERF75 was amplified using primers listed in Supplemental Table S1. Protein sequences of ERF genes used in phylogenetic tree construction and multiple sequence alignment were downloaded from the TAIR database and the NCBI database; accession numbers for each gene are listed in Supplemental Table S2 and S3. MEGA 7.0 software was used for sequence alignments and phylogenic trees. The DRE motif (A/TCCGAC)[4,5], REV motif (CAACA)[27] and CE-box motif (CCACC)[28] were manually searched. Mutations in the promoters of VvPAO1 and VvACS5 was conducted using QuikChange Lightning Site-Directed Mutagenesis Kit to generate the VvPAOm and VvACS5m constructs.
Subcellular localization and transcriptional activation analysis of VvERF75
-
Subcellular localization and transcriptional activation assays were conducted as described[29]. For these experiments, the CDS of VvERF75 without its stop codon was introduced into the pSAK277-GFP and pGBKT7 vectors, respectively. The empty vectors of pSAK277-GFP and pGBKT7 was used as controls.
Yeast one-hybrid analysis
-
The CDS sequence of VvERF75 was introduced into the pB42AD vector, while the promoters of various CCG genes were introduced into the placZi vector. Yeast one-hybrid experiments were performed as previous reported[4,30]. To accurately identify the DNA regions/motifs than can interact with VvERF75, the target genes identified in the first round of screening were further divided into 100-bp and 50-bp segments to conduct second- and third-round screens.
Dual-luciferase assays
-
For dual-luciferase assays, the promoter sequences of VvPAO1 and VvACS5 were inserted into pGreen 0800-LUC to yield reporter vectors. The CDS of VvERF75 was inserted into pSAK277 to yield effector vector. The reporter vectors and effector vectors were co-transformed into Nicotiana benthamiana leaves, with the empty pSAK277 vector used as a negative control. The experiment was carried out according to previous reports[31, 32].
EMSA assays
-
For EMSA assays, the CDS of VvERF75 was introduced upstream of the GST-tag in the pCold plasmid. Recombinant GST-VvERF75 protein was expressed in Escherichia coli strain BL21 and purified. The probe and mutated probe sequences were synthesized in Sangon Biotechnology (Shanghai, China). Each probe was biotin-labeled at the 5′ end. The reported method was used for EMSA assays[31].
Real-time quantitative PCR analysis
-
The plant RNA kit (Omega) was used for RNA extraction from frozen grape berry samples. qRT-PCR was carried out according to a previous report[32] with GAPDH (CB973647) serving as an internal control using the primers listed in Supplemental Table S1.
Genetic transformation and confirmation of VvERF75
-
The CDS sequence of VvERF75 was inserted into the binary vector pSAK277 to generate the 35Spro-VvERF75-GFP construct. Tomato (Solanum lycopersicum cv. Micro-Tom) plants were used for VvERF75 overexpression. Transgenic plants were generated and cultured as described[30]. DNA and RNA were extracted from leaves of candidate transgenic plants to identify VvERF75 overexpressing (VvERF75-OE) plants by qRT-PCR and PCR. The expression of SlPAO, SlACS2 and SlACS4 in tomato were detected by qRT-PCR using primers in Supplemental Table S1.
Measurement of ethylene production, chlorophyll and lycopene content in fruit and rachis of tomato
-
Fruits of VvERF75-OE transgenic and WT tomato plants were detached at the breaker, breaker+3d, breaker+5d, and breaker+7d stages of fruit ripening. Ethylene production, and chlorophyll level content were measured as described previously[33−35].
Statistical analysis
-
Experiments in this study were conducted in three replicates. Statistics were analyzed using DPS software. Error bars represent standard deviation. * indicate significant difference (P < 0.05) as obtained by one-way analysis of variance and the least significant difference test.
-
Ethylene treatment of maturing grape clusters was carried out one week before veraison, and clusters were detached 5, 10 and 15 d after treatment (DAT). Berry coloration was advanced by about 5 d in ethylene-treated clusters compared with controls (Fig. 1a). Single berry weight (Fig. 1b), longitudinal diameter (Fig. 1c) and transverse diameter (Fig. 1d) showed no significant variation after treatment, while total soluble solids were higher at 5 d after treatment but similar to the controls afterwards (Fig. 1e). Titratable acid (Fig. 1f) and chlorophyll content (Fig. 1g) were decreased, while anthocyanin content (Fig. 1h) and ethylene production (Fig. 1i) were increased after ethylene treatment (Fig. 1a). This indicated that ethylene treatment can promote coloration of ‘Summer Black’ grape.

Figure 1.
The effects of ethylene treatment on fruit ripening in ‘Summer Black’ grape. (a) Ripening phenotype in response to ethylene treatment (on day 0), Bar = 0.5 cm. (b) Single berry weight. (c) Longitudinal diameter. (d) Transverse diameter. (e) Total soluble solids. (f) Titratable acid. (g) Anthocyanin content. (h) Chlorophyll content. (i) Ethylene production.
VvERF75 belongs to the ERF IXa subfamily and has a AP2/ERF domain
-
ERF transcription factors take part in fruit ripening by modulating expression of genes related to ethylene production or chlorophyll degradation[2,3,36]. The ERF transcription factor family can be classified into 12 subfamilies[20]. We identified VvERF75 in our previous genome analysis of genes responsive to 1-MCP treatment[34]. Here, VvERF75 expression was negatively correlated with chlorophyll content, with a Pearson correlation coefficient of 0.799* (Supplemental Fig. S1) . A phylogenetic tree of VvERF75 and some of the ERF proteins from Arabidopsis revealed that VvERF75 belongs to subfamily IX (Supplemental Fig. S2a). Group IX can be further divided into Subgroups IXa, IXb and IXc, with VvERF75 belonging to group IXa (Supplemental Fig. S2b). The Chl degradation and ethylene biosynthetic roles of most genes in group IXa are largely unclear. Alignment of protein sequences of VvERF75 and its homologs from Ziziphus jujube (ZjPIT75-like), Mangifera indica (MiPIT5-like), Quercus suber (QsPIT5-like), Citrus sinensis (CsPITS) and Medicago truncatula (MtPIT5) showed that VvERF75 contains a conserved AP2/ERF domain (ß-1 ß-2, ß-3, and α-helix) (Supplemental Fig. S2c).
The expression of the transcription factor VvERF75 and several potential downstream CCGs (VvPAO1, VvPAO2, VvNYC1, VvNOL, VvCLH1 and VvRCCR) were examined after veraison. Expression of VvERF75 and VvPAO1, VvPAO2, VvCLH1 and VvNOL increased in response to ethylene treatment, while expression of VvCLH2 showed more variation, decreasing at 5 DAT and increasing at 10 DAT (Fig. 2). Expression of VvERF75 showed a similar pattern to ethylene production (Fig. 1i) and expression of the chlorophyll degradation genes VvPAO1, VvPAO2, VvCLH1, VvNOL and VvCLH2 (Fig. 2), but the opposite pattern to chlorophyll content in grape after ethylene treatment (Fig. 1g). These correlations indicated that VvERF75 may have a potential role in regulation of ethylene biosynthesis and chlorophyll degradation.
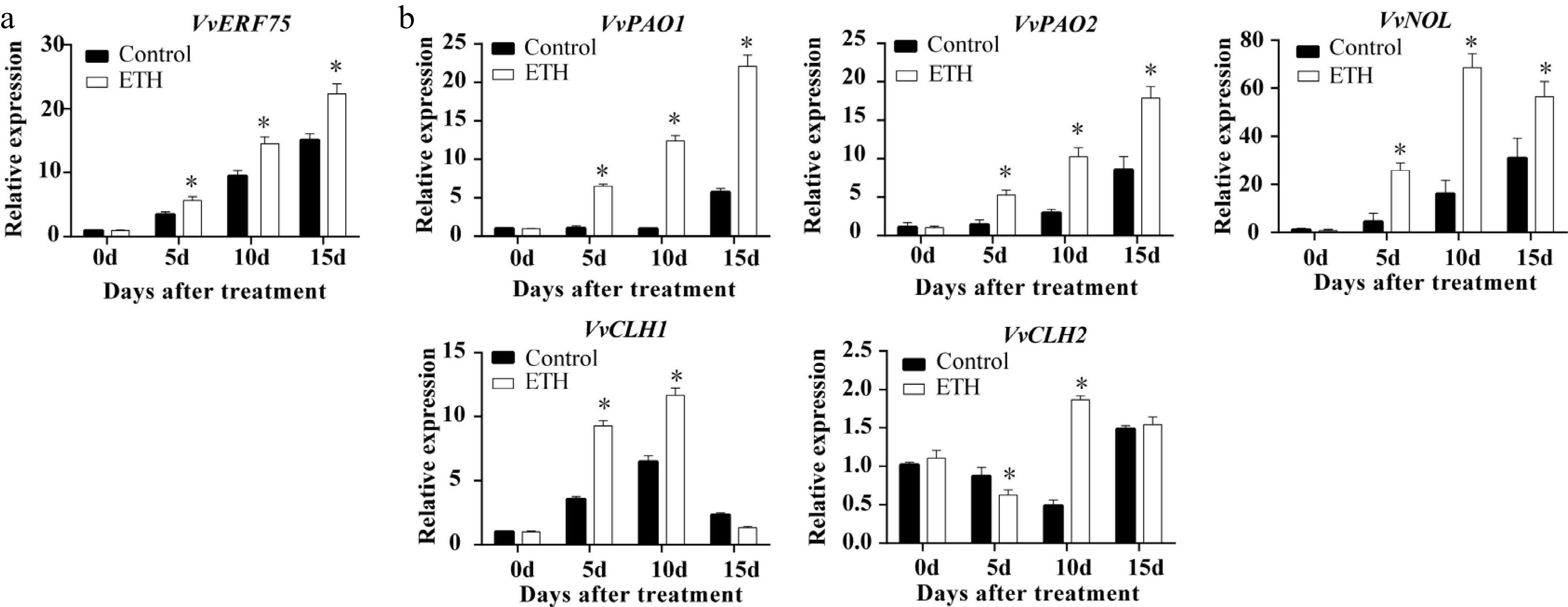
Figure 2.
Expression in ‘Summer Black’ grape of (a) VvERF75 and (b) chlorophyll degradation related genes in response to ethylene treatment.
VvERF75 is localized in the nucleus and has transcriptional activity
-
The GFP-VvERF75 construct was transiently transformed into tobacco leaves to verify the subcellular localization of VvERF75. The fluorescence of GFP alone was observed in both the nucleus and the cytoplasm. The fluorescence of the GFP-VvERF75 protein was observed only in the nucleus, as verified with NLS-mCherry (a nuclear-localized marker) (Fig. 3a), suggesting that VvERF75 is localized in the nucleus.
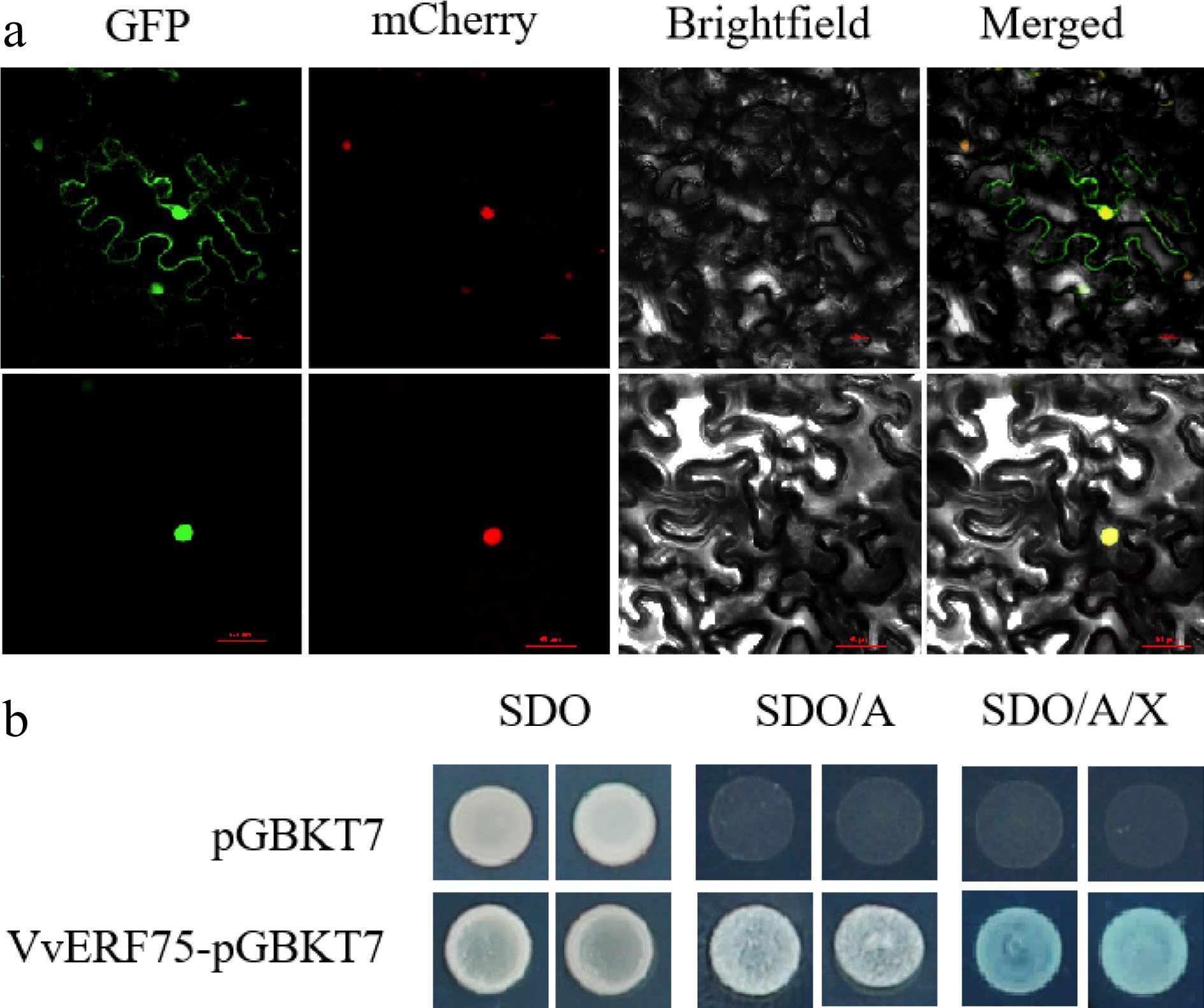
Figure 3.
Subcellular localization in tobacco and transcriptional activation in yeast of VvERF75. (a) Subcellular localization of VvERF75 in Nicotiana benthamiana leaves. VvERF75-GFP was transformed into Nicotiana benthamiana. VvERF75-GFP, GFP signal; Brightfield, white light; Merged, combined GFP and brightfield signals. NLS mCherry was used as a nuclear-localized marker. Bar = 50 μm. (b) VvERF75 trans-activation activity assay. Yeast cells were dotted on plates containing either SDO, synthetic dropout media; SDO/A, synthetic dropout medium supplemented with Aureobasidin A but lacking tryptophan; or SD/AbA/X, synthetic dropout medium supplemented with Aureobasidin A and X-a-Gal but lacking tryptophan. pGBKT7 vector was used as negative control.
A transcription activation assay was conducted in yeast to identify if VvERF75 has transcriptional activity. VvERF75-pGBKT7 and the empty pGBKT7 vector were separately transformed into yeast strain Y2H gold. All transformants survived on synthetic drop-out media (SDO) media. Yeast transformants with VvERF75 grew successfully in the selective media SDO/A and SDO/A/X, while yeast cells transformed with pGBKT7 vector did not (Fig. 3b). These results provide evidence that VvERF75 activated transcription in yeast cells.
VvERF75 activated expression of VvPAO1 and VvACS5
-
Yeast one-hybrid and luciferase assays were performed to detect any relationship between VvERF75 and Chl degradation-related genes. The results suggest that VvERF75 can bind to the promoter of VvPAO1 (Fig. 4a) and positively regulate its expression (Fig. 4c).
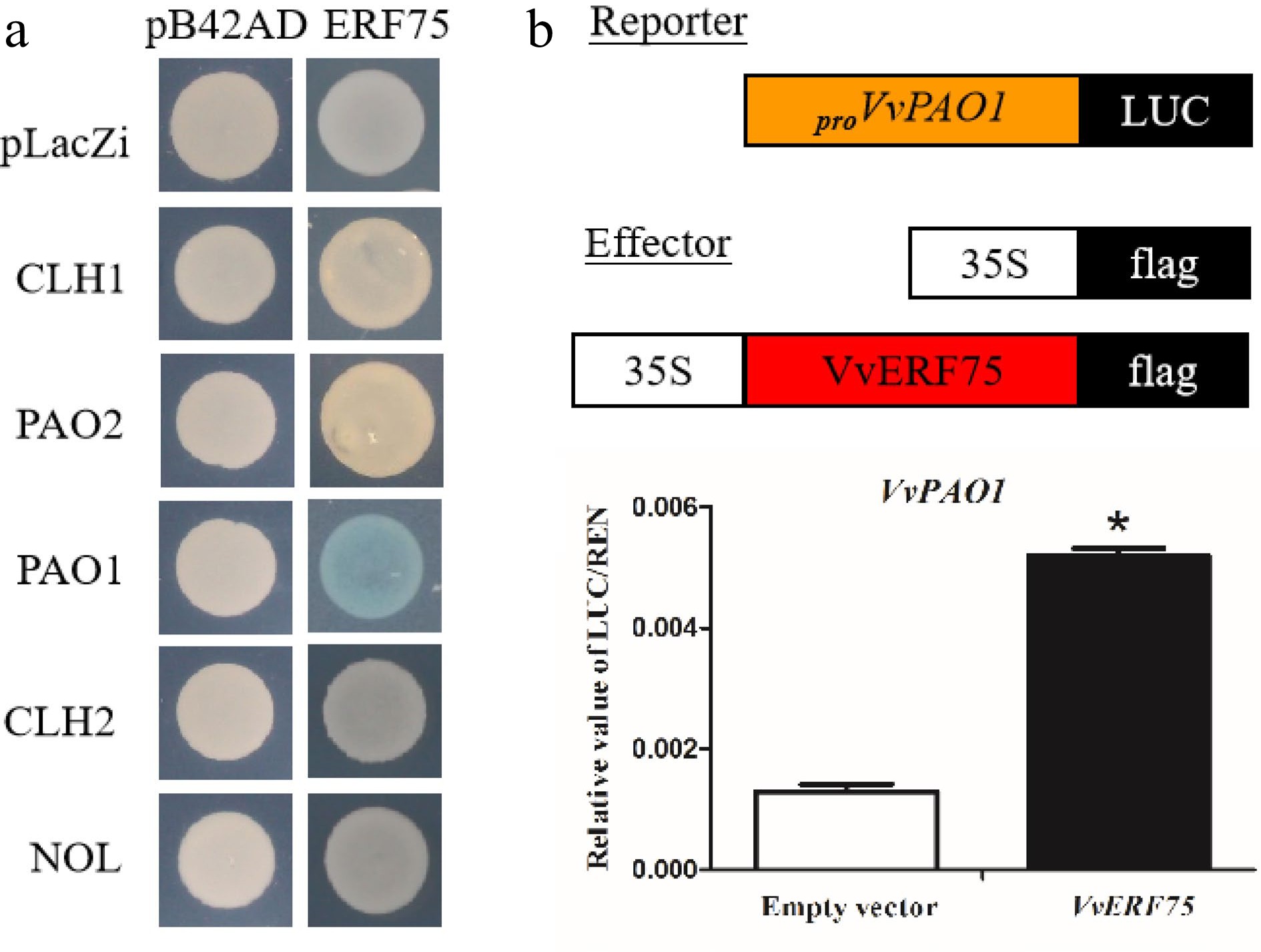
Figure 4.
Analysis of VvERF75 regulation of VvPAO1. (a) Yeast one-hybrid analysis of VvERF75 binding to the promoters of chlorophyll catabolic genes. (b) Schematics of the reporter and effector constructs used in dual-luciferase reporter assay. (c) Activation of the VvPAO1 gene by VvERF75 is indicated by the LUC/REN ratio. Empty vector: 35S:flag + proVvPAO:LUC; VvERF75: 35S:VvERF75:flag + proVvPAO:LUC, the empty effector was used as a control.
To identify specific VvERF75 binding sites in the VvPAO1 promoter sequence, a sequential deletion series was generated and assessed via yeast one-hybrid assays and EMSA. The fragment p5 contains the DRE motif. The results indicated that VvERF75 can bind to the DRE motif in the promoter of VvPAO1 (Fig. 5a & b). Mutation of the DRE motif in the fragment p5-3 disrupted the interaction between VvERF75 and the PAO1 fragment in yeast (Fig. 5c). This interaction was further confirmed by EMSA (Fig. 5d), in which the VvERF75-GST fusion protein bound directly to the promoter of VvPAO1 (Fig. 5d), but failed to bind to the mutant DRE motif of VvPAO1 (Fig. 5d).
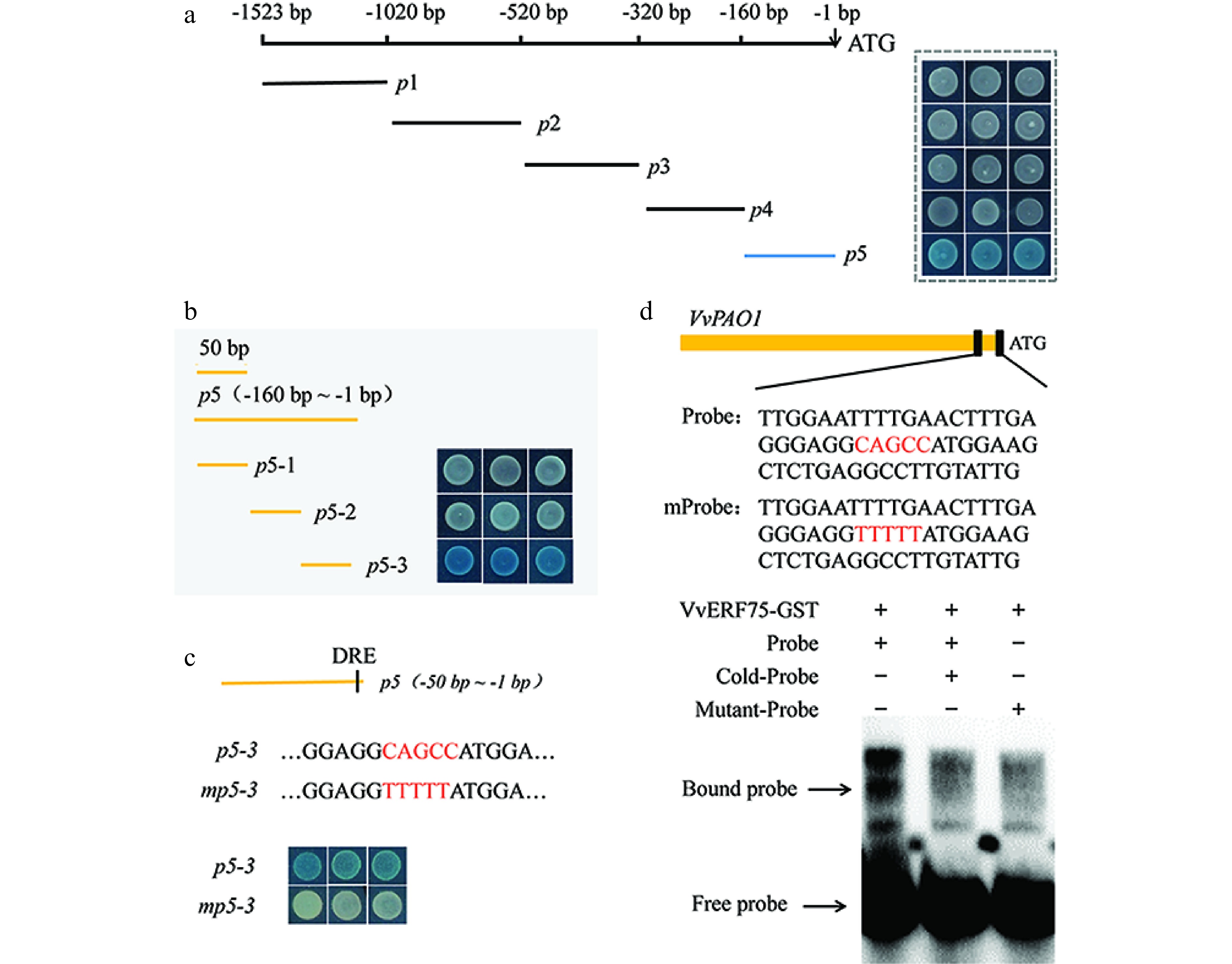
Figure 5.
Electromobility shift and yeast one-hybrid assays showing the binding of VvERF75 to the promoter of VvPAO1. (a), (b) Sequential deletion series and (c) mutation of VvPAO1 promoter for use in yeast one-hybrid assays with VvERF75. (d) Sequence of the probe derived from the promoter of VvPAO1 (p5-3) and VvPAO1m are shown, with the DRE motif and its mutant indicated in red. (e) EMSA of VvERF75 protein binding to the promoter sequence of the VvPAO1 gene. + and − represent the presence or absence of the respective composition in the binding experiment. The labeled 'free probe' and DNA protein complex 'bound probe' positions are indicated, showing competition with an unlabeled fragment for each promoter domain.
ERF transcription factors have been reported to regulate the expression of ACS or ACO, and thus, regulate ethylene biosynthesis. The increased production of ethylene in grape berries raises the question of whether VvERF75 can modulate ethylene biosynthesis during berry ripening. VvACS5 has a DRE/GCC-box motif and is differentially expressed during rachis browning[36], so was selected for further analysis. Yeast one-hybrid and dual-luciferase experiments (Fig. 6b) revealed that VvERF75 can bind to the promoter of VvACS5 and positively regulate its expression (Fig. 6a & c). The DRE and GCC motifs in the promoter of VvACS5 were mutated (m1 and m2), and binding of VvERF75 on these promoter sequences was assessed by dual-LUC experiments. The LUC/REN expression ratio in the m1 and m2 variants was not significantly different with that of the empty vector, which implied that VvERF75 binds the intact DRE motif in the promoter of VvACS5 (Fig. 6d). In the EMSA, the VvERF75-GST fusion protein can only bind directly to the DRE motif of VvACS5 (Fig. 6e), but not the GCC-box motif of VvACS5 (Fig. 6f). Taken together, these results indicated that VvERF75 might regulate ethylene biosynthesis and Chl degradation genes during color development in grape berry.
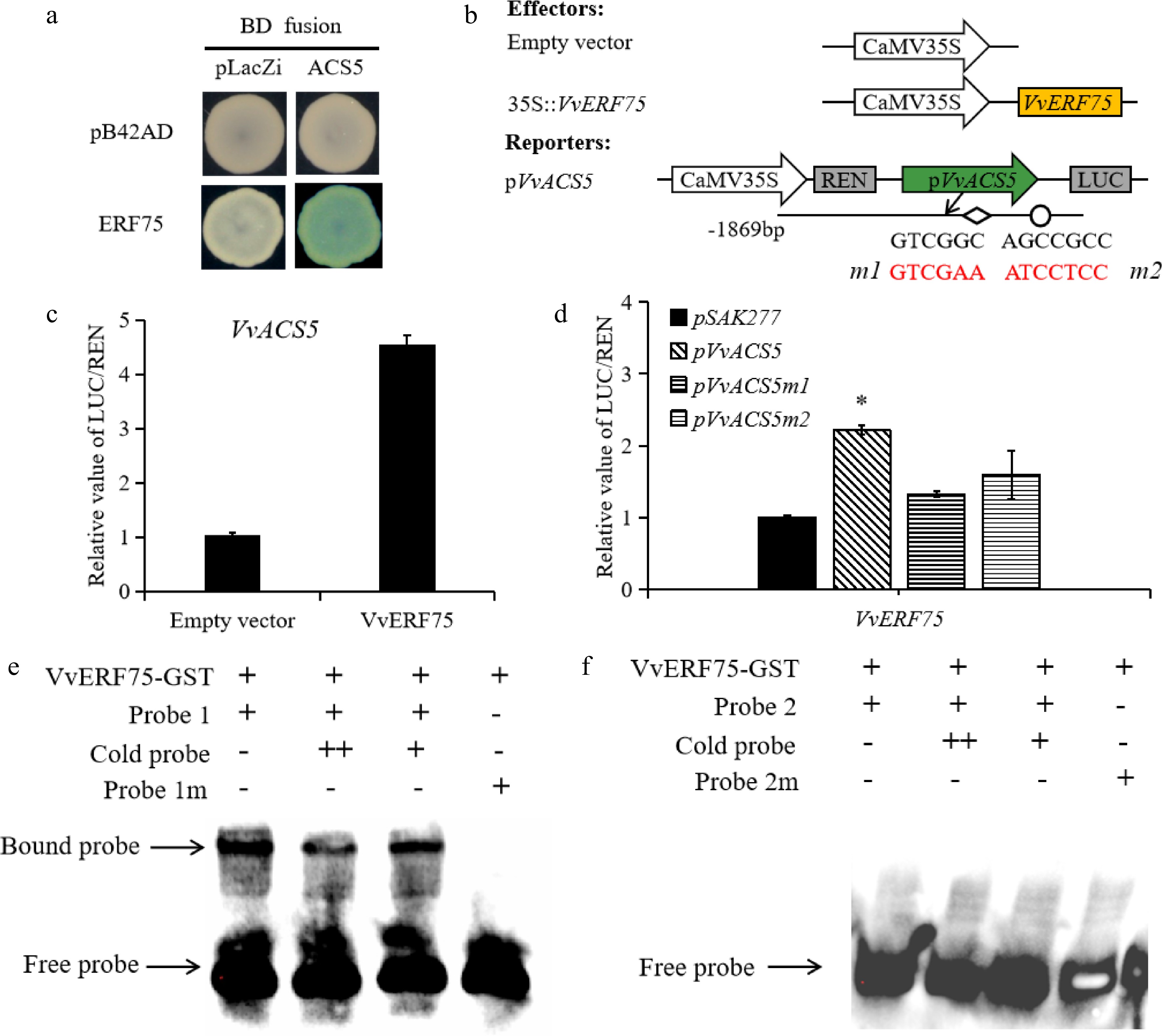
Figure 6.
Regulation of VvACS5 by VvERF75. (a) Yeast one-hybrid assay analysis of VvERF75 binding to VvACS5 promoter. (b) Schematics of the reporter and effector constructs used in the dual-luciferase reporter assay, the DRE motif and its mutant indicated in red. (c) Dual-luciferase assays of VvERF75 on promoter of VvACS5. (d) VvERF75 activation of the wild-type and mutant promoters of the VvACS5 genes. m1 carries a mutation of the DRE motif and m2 of the GCC box in the promoter of VvACS5. (e), (f) EMSA of VvERF75 protein binding to motifs 1 and 2 on promoter of the VvACS51 gene. + and − represent the presence or absence of the respective component in the binding experiment. The labeled 'free probe' and DNA protein complex 'bound probe' positions are indicated, showing competition with an unlabeled fragment for each promoter domain.
VvERF75 affects flowering time and plant height in transgenic tomato
-
To investigate the function of VvERF75 during plant development, VvREF75 was overexpressed in tomato via Agrobacterium-mediated transformation. VvERF75-OE plants were identified by PCR and qRT-PCR (Fig. 7a & b). Three independent lines (75-4, 75-7 and 75-8) that showed higher expression levels of VvERF75 were selected for further analysis and taken to the T1 generation (Fig. 7c). Flowering time of the VvERF75-OE transgenic plants was advanced by about 2-3 d compared with WT, which flowered at an average of 43 d after sowing (Fig. 7d). In addition, the height of the VvERF75-OE plants was significantly decreased compared to the WT plants (Fig. 7e). Our results implied that VvERF75 plays a critical role in plant development.
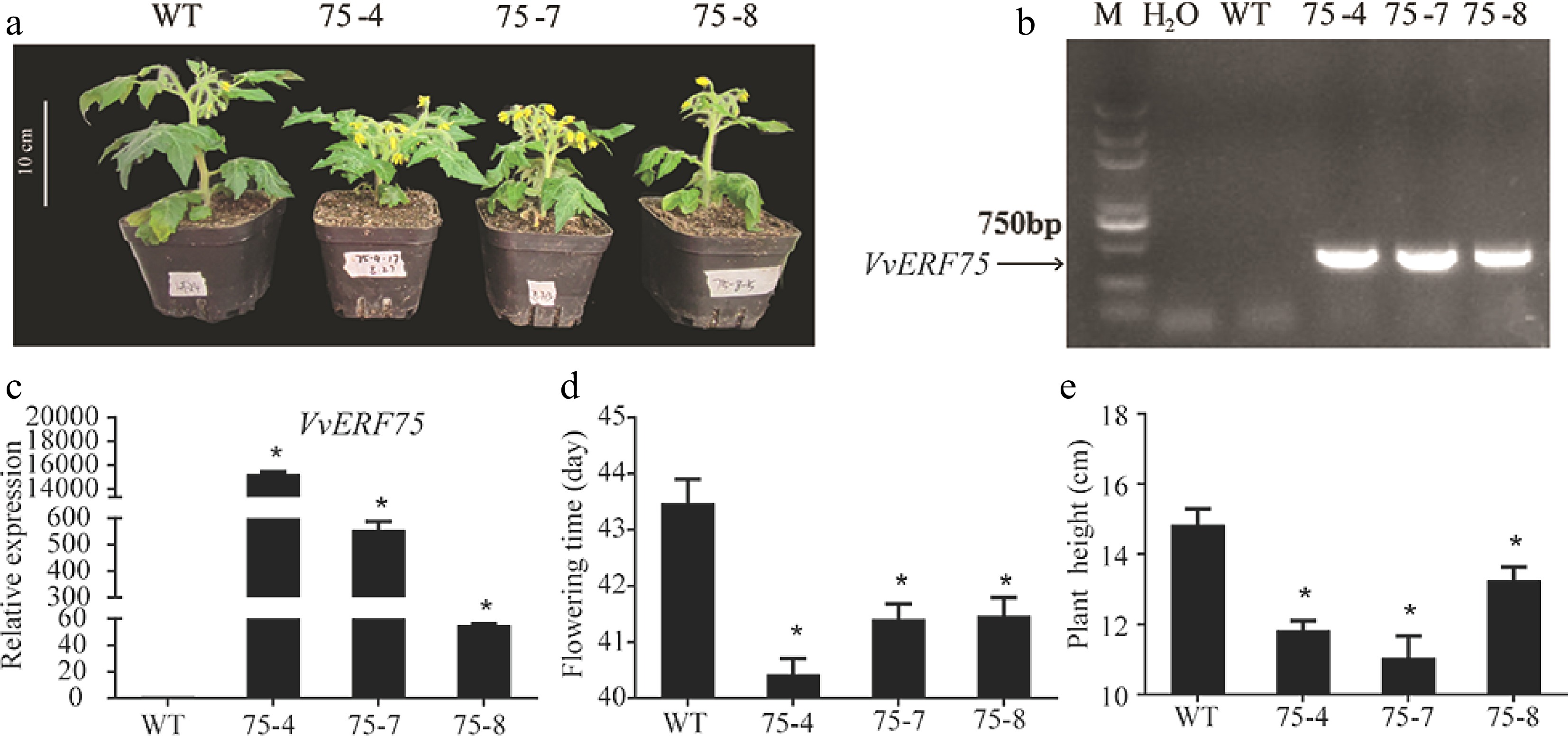
Figure 7.
Overexpression of VvERF75 in tomato changed flowering time and plant height. (a) Transgenic plants of VvERF75. Detection of transgenic VvERF75 plants by (b) PCR and (c) qRT-PCR. (d) Flowering time of transgenic VvERF75-OE plants compared to WT. (e) Height of transgenic VvERF75-OE plants compared to WT.
VvERF75 enhanced ethylene production and chlorophyll degradation in transgenic tomato
-
Fruits reached the breaker (Br) stage 42 d post-anthesis (DPA) in WT plants (Fig. 8a), but the VvERF75-OE lines began to turn red about 3 d later. Ethylene production, Chl and lycopene contents were monitored from Br to Br+7. In WT plants, ethylene production and lycopene contents increased while Chl content decreased during fruit ripening (Fig. 8b−d). In the VvERF75-OE plants, the ethylene production was higher than in WT (Fig. 8b), while the Chl content was lower in the VvERF75-OE lines (Fig. 8c) and the lycopene content was increased (Fig. 8d).
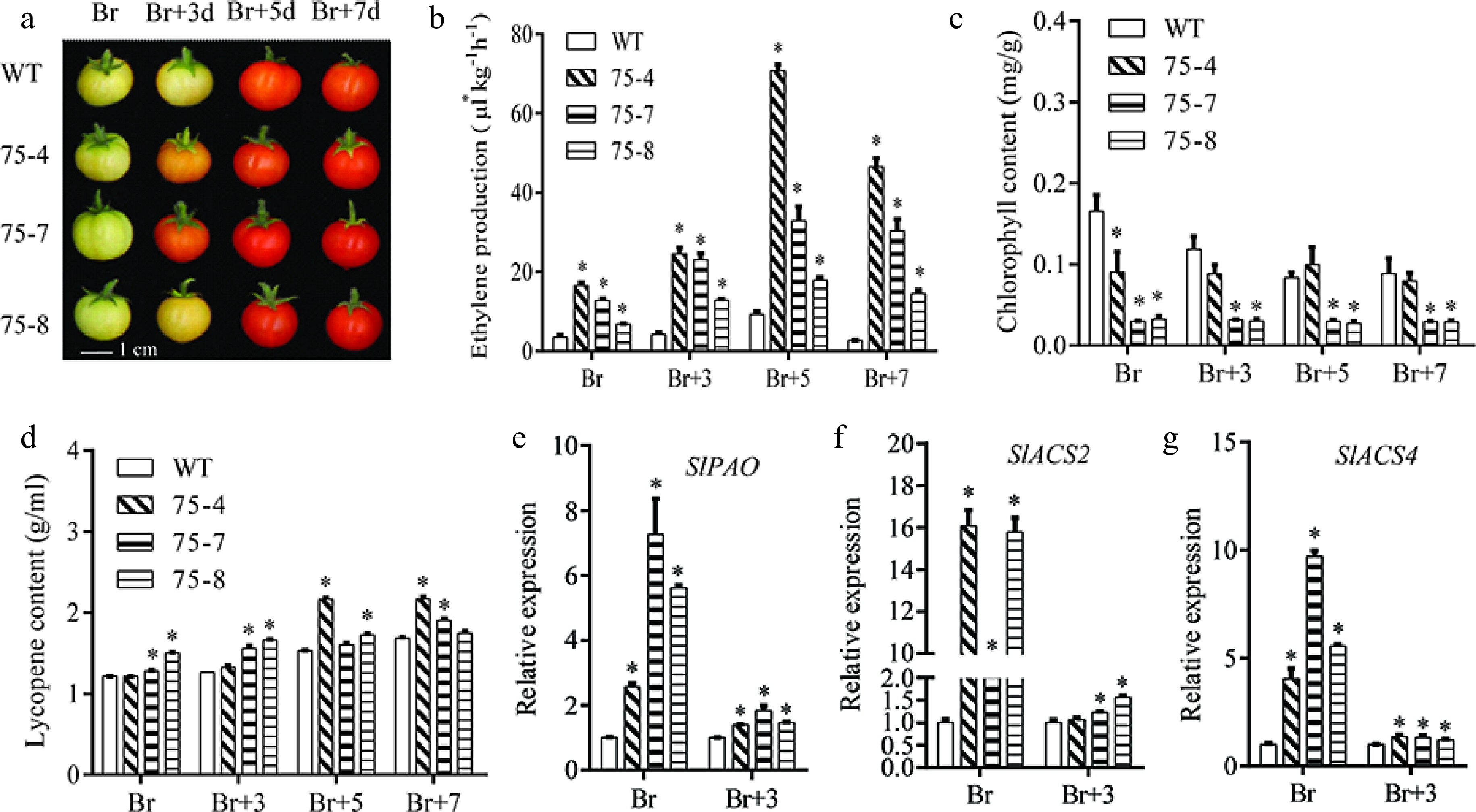
Figure 8.
VvERF75 enhanced ethylene production and chlorophyll degradation in tomato. (a) Color phenotype of VvERF75 overexpression lines. (b) Ethylene production, (c) chlorophyll content, (d) lycopene content in WT and VvERF75 overexpression lines at Br, Br+3, Br+5 and Br+7 stage. Expression of (e) SlPAO, (f) SlACS2 and (g) SlACS4 in WT and VvERF75-overexpression lines at Br and Br+3 stage.
Tomato fruits at the Br and Br+3 stage, which showed obvious differences, were selected for gene expression analysis. Expression levels of SlPAO, SlACS2 and SlACS4 were significantly increased in VvERF75-OE plants compared with WT (Fig. 8e−g). Together these data further support that VvERF75 can regulate expression of PAO and ACS, thus, modulating chlorophyll degradation and ethylene biosynthesis.
-
Ethylene has been proven to play vital roles during plant development[37−39] and to control downstream transcriptional regulators, the ERFs, that are responsible for the ethylene signal. ERFs are in the AP2/ERF superfamily and carry the AP2/ERF domain, which is 60-70 amino acids long and binds DNA. The superfamily can be further divided into the AP2 family, the ERF family, and the RAV family[20]. The ERF family in Arabidopsis can be classified into 12 subfamilies[20]. The ERF IX subfamily can divide into three subgroups, IXa, IXb and IXc[20], phylogenetic analysis and sequence alignment revealed that VvERF75 belongs to subfamily IXa (Supplemental Fig. S2a & Sb) and contains an AP2/ERF conserved domain (Supplemental Fig. S2c). ERF transcription factors can recognize and bind to the GCC-box/DRE motifs in promoters of downstream genes to activate their expression[2,3,5,40]. Genes of the ERF IX subfamily participate in resistance to abiotic stress[41−43], cold stress[44,45], and ethylene synthesis and response[3,46,47]. There are 40 predicted proteins in the grape ERF IX subfamily[25], some of which have been functionally characterized. VaERF092 from Amur grape regulates cold tolerance[48], VaERF20 enhances resistance in tomato to Pseudomonas syringae pathovar tomato (Pst) DC3000[49]. Our previous research revealed that the Chl content was reduced during rachis degreening and that VvERF95 could modulate Chl degradation in this process[35]. Nevertheless, little is known about any roles of genes in the grape ERF IX family during berry ripening.
Ethylene and Chl degradation have been proved to be main aspects of berry coloration during grape fruit ripening[50,51], but the mechanism underlying chlorophyll degradation during fruit ripening remains largely unknown. Our previous report revealed that VvERF95 can activate expression of VvPAO1, thus accelerating Chl degradation during rachis browning, and that the motifs for ERF binding exist in the promoters of many CCGs[35]. For example, CitERF6 can activate expression of CitPPH to promote Chl degradation during citrus fruit degreening[40]; VvERF17 regulates the expression of VvNOL, VvPPH, VvPAO and VvRCCR to mediate Chl degradation in grape berry coloration[40]; apple ERF17 binds the promoter of PPH and NYC to regulate Chl degradation during fruit peel degreening[4,52]. In this study, expression of VvERF75 increased during grape berry ripening in response to ethylene treatment, while Chl content decreased, ethylene release increased, and the transcript levels of Chl degradation and ethylene synthesis genes increased. Our data demonstrated that VvERF75 can activate expression of VvPAO1 (Fig. 3 & 4), which is like the activity of VvERF95[35].
In addition, ERF proteins have been shown to modulate ethylene in banana and apple by regulating the expression of ACO or ACS[3,23]. Our experiment revealed that VvERF75 can modulate expression of VvACS5. Site mutation of the DRE motifs revealed that VvERF75 can bind to the promoters of VvPAO1 (Fig. 4c & d) and VvACS5 (Fig. 5d) at the DRE motifs, which is consistent with previous reports[2,3,5,40].
To further characterize the role of VvERF75 in chlorophyll degradation and ethylene production, VvERF75 was transformed into tomato. Overexpression of VvERF75 in tomato resulted in decreased content of Chl and increased production of ethylene during fruit development as well as the increased expression of SlPAO, SlACS2 and SlACS4, further validating that VvERF75 activates the expression of PAO1 and ACS5. These results are consistent with results for VvERF17[26] and VvERF95[35] in grape as well as ERFs in apple and banana[3,23], revealing that the ability of ERF transcription factors to regulate chlorophyll degradation and ethylene biosynthesis is conserved among different species.
-
In brief, our findings reveal that VvERF75 belongs to the ERF IXa subfamily, has an AP2 domain, is localized in the nucleus, and has transactivation activity in yeast cells. During grape berry coloration, expression of VvERF75 increased, activating the expression of VvPAO1 by binding to the DRE motif in its promoter. This accelerates degradation of chlorophyll. VvERF75 can also modulate ethylene biosynthesis by activating the expression of VvACS5, leading to increased ethylene production, which induces chlorophyll degradation. In addition, VvERF75-OE tomato plants showed reduced plant height and advanced flowering and fruit ripening times. Our results provide new insight into how VvERF75 impacts chlorophyll degradation and ethylene production during fruit ripening in grape. Our results also expanded the knowledge of the diverse functions of ethylene in non-climacteric fruits.
This work was supported by the Natural Science Foundation of China (32002017), the Henan Province Outstanding Foreign Scholar Program (GZS2020007), and the Natural Science Foundation of Henan Province (222300420457).
-
The authors declare that they have no conflict of interest.
-
# These authors contributed equally: Zhiqian Li, Chaoyang Chen, Dongfang Zou
- Supplemental Fig. S1 Correlation between expression of VvERF75 and content of chlorophyll during grape berry development.
- Supplemental Fig. S2 Sequence analysis of VvERF75. (A) Phylogenetic analysis of VvERF75 and other proteins in the ERF family that represent each of the 12 subfamilies in Arabidopsis thaliana. (B) Phylogenetic analysis of VvERF75 and the ERF IX subfamily proteins in Arabidopsis thaliana. The phylogenic trees were constructed using MEGA 7 (neighbor-joining method, 1000 bootstraps using). The red circle indicates VvERF75. (C) Multiple amino acid sequence alignment of VvERF75 and homologous proteins. The alignments were performed using ClustalX. Conserved amino acid residues were denoted by black and light gray shading. The AP2/ERF domain is underlined with a black line. Predicted α-helix and β-sheet regions were represented with black bar and arrowheads. The amino acid sequences were obtained from The Arabidopsis Information Resource (TAIR) database and National Center for Biotechnology Information (NCBI) database.
- Supplemental Table S1 Primers used in this study.
- Supplemental Table S2 Gene accession number used in Figure 3a and b.
- Supplemental Table S3 Gene accession number used in Figure 3c.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Li Z, Chen C, Zou D, Li J, Huang Y, et al. 2023. Ethylene accelerates grape ripening via increasing VvERF75-induced ethylene synthesis and chlorophyll degradation. Fruit Research 3:3 doi: 10.48130/FruRes-2023-0003
Ethylene accelerates grape ripening via increasing VvERF75-induced ethylene synthesis and chlorophyll degradation
- Received: 20 November 2022
- Accepted: 06 February 2023
- Published online: 24 February 2023
Abstract: Ethylene plays an important role in grape ripening, acting through ethylene response factors (ERFs) that are downstream of ethylene signaling pathways, yet little else is known about these molecular mechanisms. In our study, we demonstrate that ethylene treatment promotes grape ripening and that the transcription factor VvERF75 shows increased expression during grape ripening. VvERF75 was found to contain a conserved AP2/ERF domain, to be a member of the ERF IXa subfamily, and to localize to the nucleus and to have transactivation activity in yeast. Our in vitro assays showed that VvERF75 can positively regulate expression of pheophorbide a oxygenase (VvPAO1) and 1-aminocyclopropane-1-carboxylic acid synthase (VvACS5) by binding to DRE motifs in their promoters. Overexpression of VvERF75 in tomato increased ethylene production and resulted in dwarfed plants, early flowering and fruit ripening, increased SlPAO and SlACS transcript levels, and decreased chlorophyll content. In general, our study revealed that VvERF75 may regulate fruit ripening by promoting ethylene biosynthesis and chlorophyll degradation, a discovery that lays a foundation for deciphering the molecular mechanisms of grape ripening.













