-
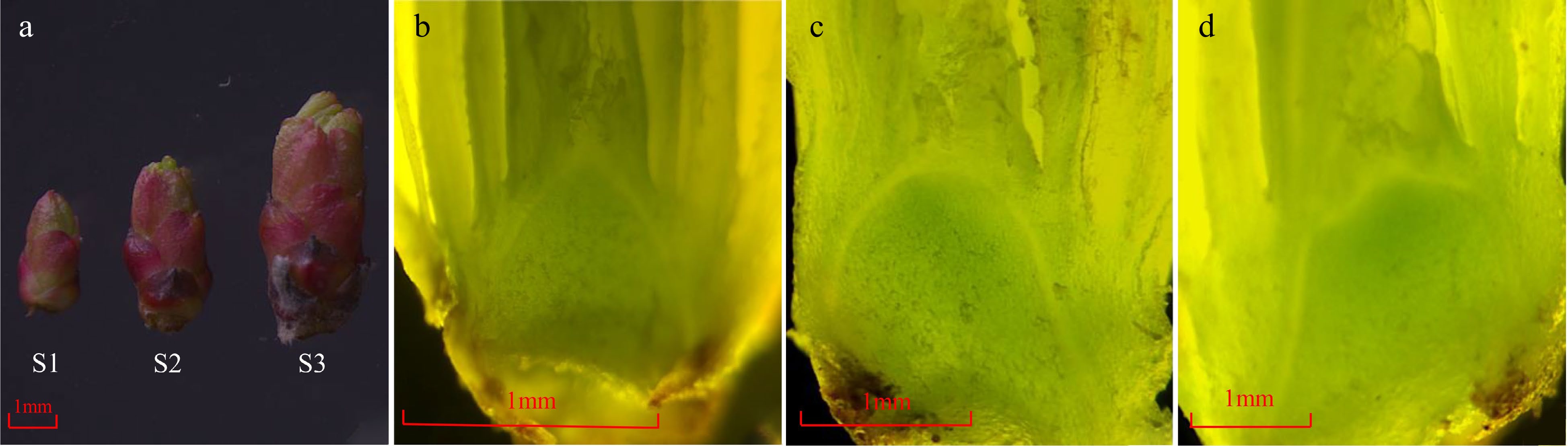
Figure 1.
The morphology changes of buds during the flowering transition process in R. rugosa ‘Duoji Huangmei’. (a) The external morphology of buds at different development stages. (b) The growth cone was pointed conical at S1. (c) The growth cone was oval at S2. (d) The sepal primordium began to bulge at S3.
-
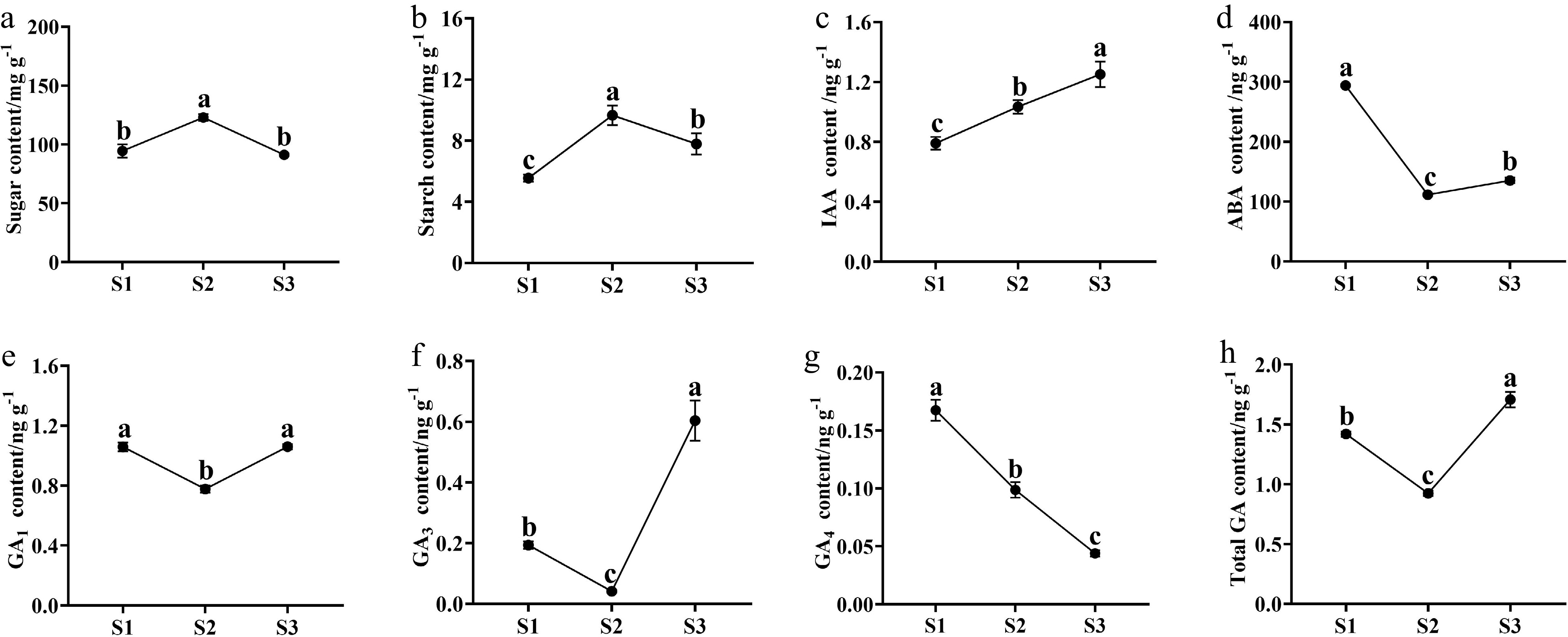
Figure 2.
The content changes of sugars, starch, and endogenous phytohormones in buds during the flowering transition process of R. rugosa 'Duoji Huangmei'. (a) Sugar content. (b) Starch content. (c) IAA content. (d) ABA content. (e) GA1 content. (f) GA3 content. (g) GA4 content. (h) Total GA content. The different small letter for each content indicated significant difference between germplasms at ɑ = 0.05.
-
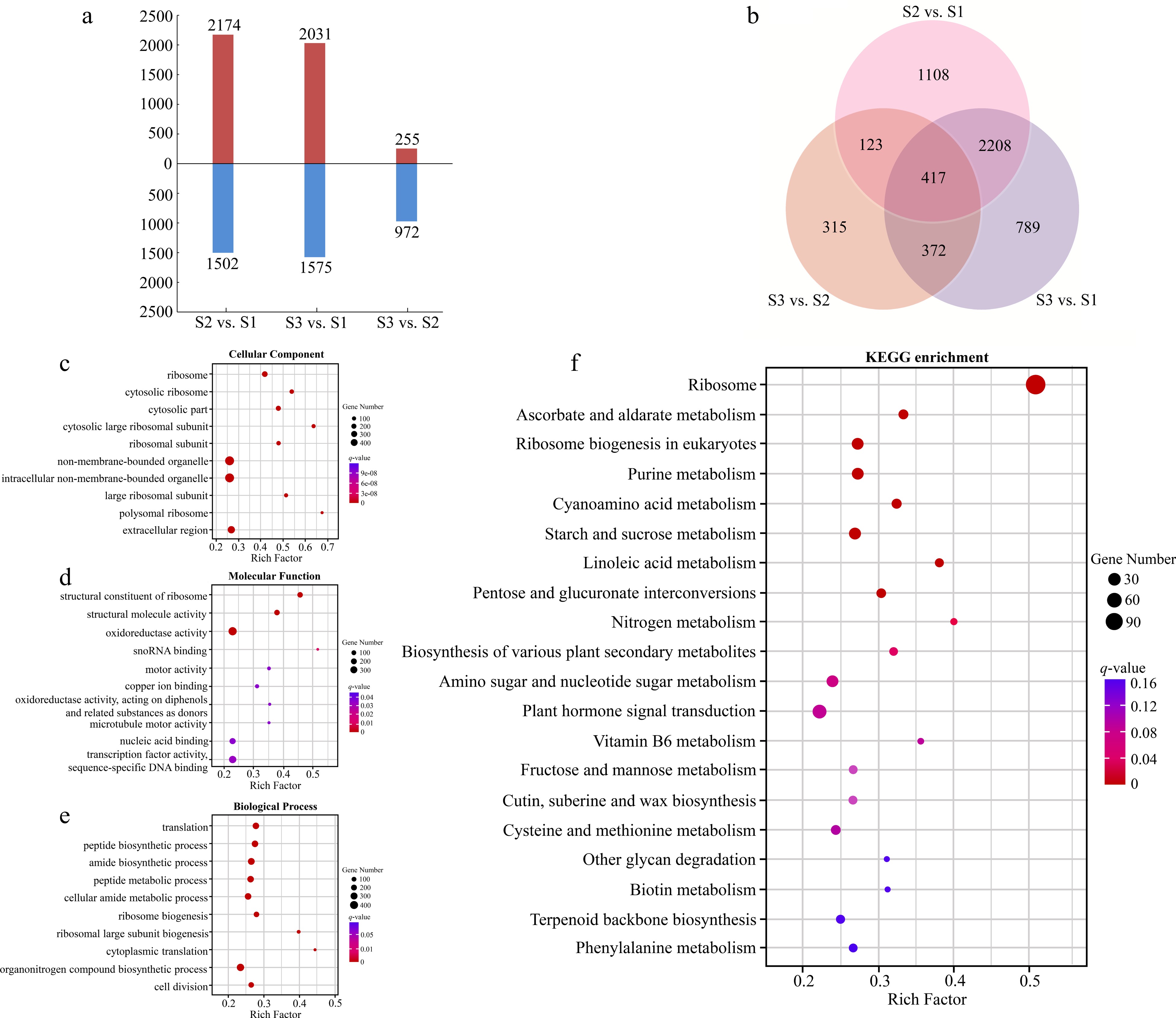
Figure 3.
Analyses of DEGs at three stages of flowering transition in R. rugosa. (a) Numbers of DEGs in each comparison. (b) Venn diagram analyses of differential and stage-specific expression genes in each comparison. (c) The GO terms of DEGs in the cellular components. (d) The GO terms of DEGs in the molecular functions. (e) The GO terms of DEGs in the biological process. (f) KEGG analysis of DEGs.
-
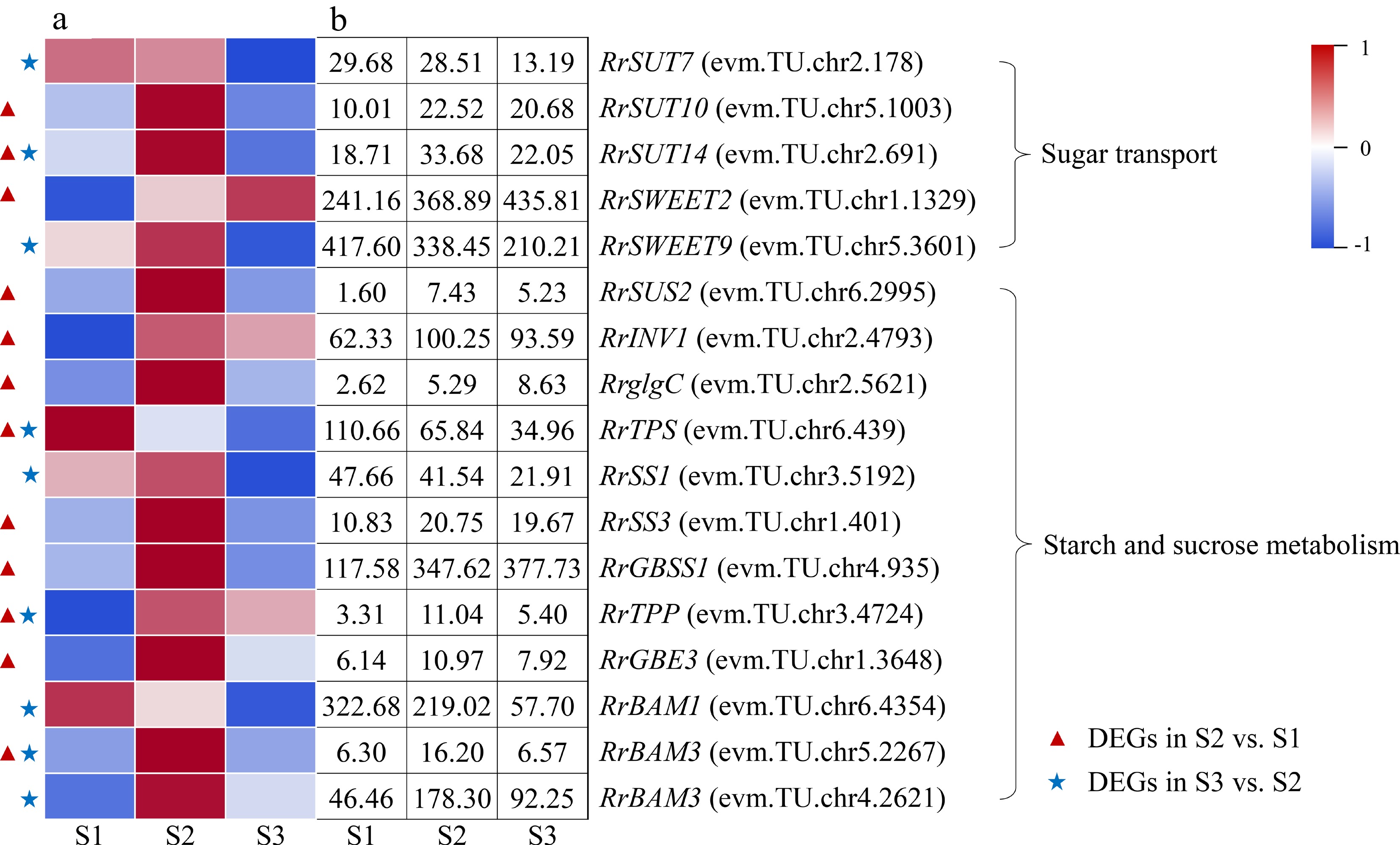
Figure 4.
Expression of starch- and sucrose-related DEGs. (a)The expression heat map of DEGs. (b) The FPKM values of DEGs. BAM: Beta-amylase; GBE3: 1,4-alpha-glucan-branching enzyme 3; GBSS1: Granule-bound starch synthase 1; glgC: Glucose-1-phosphate adenylyltransferase; INV1: insoluble isoenzyme CWINV1-like; SS1: Starch synthase 1; SUS2: Sucrose-phosphate synthase 2; SUT: Sugar transport protein; SWEET: Sugars will eventually be exported transporter; TPP: Trehalose-phosphate phosphatase; TPS: Trehalose-phosphate synthase
-
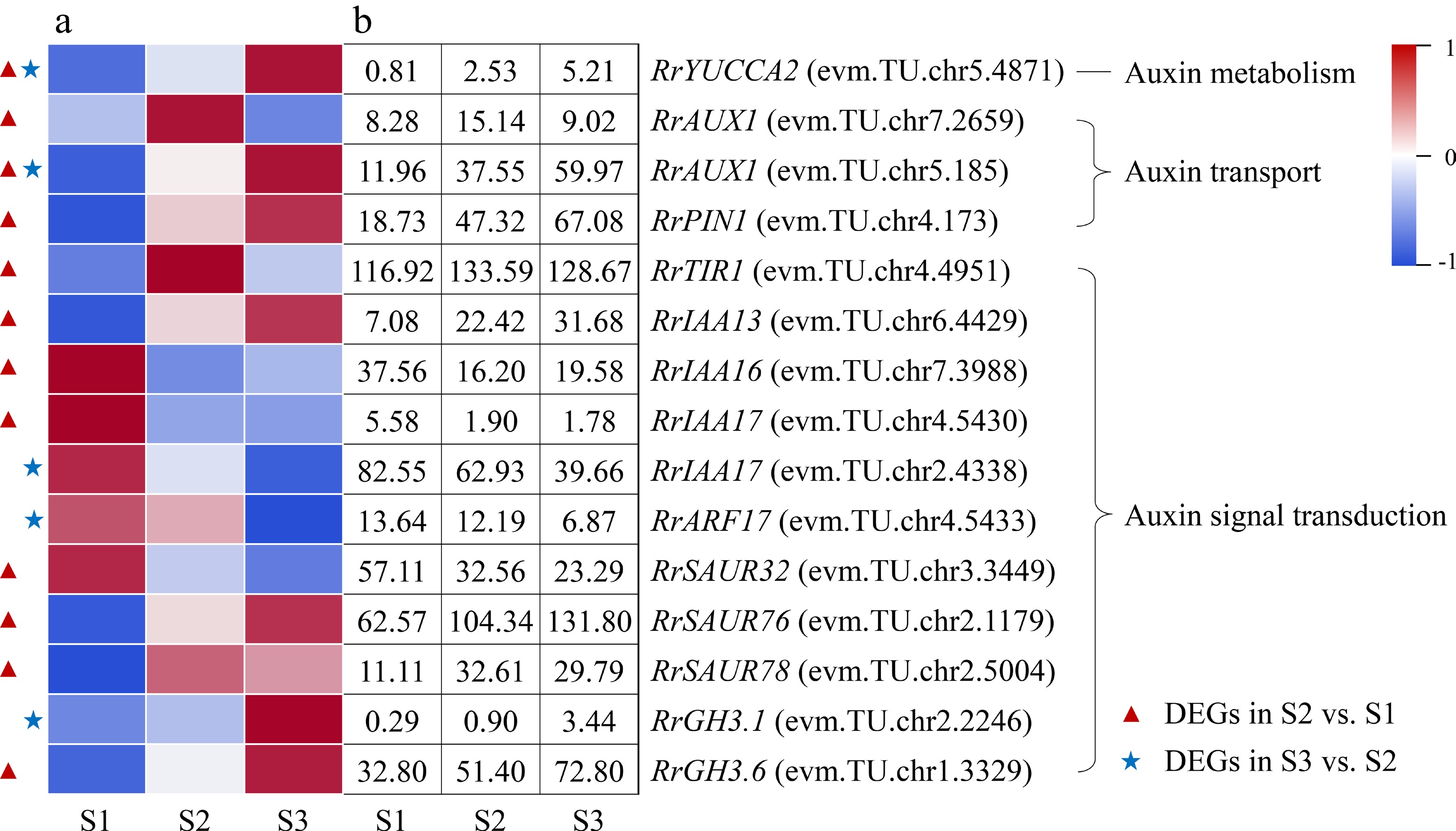
Figure 5.
Expression of IAA-related DEGs. (a) The expression heat map of DEGs. (b) The FPKM values of DEGs. ARF: Auxin response factor; AUX1: Auxin 1; GH3: Gretchen hagen 3; PIN1: PIN-formed acicular protein 1; SAUR: Small auxin-up RNA protein; TIR1: Transport inhibitor response protein 1;YUCCA2: YUCCA flavin-containing monooxygenase 2
-
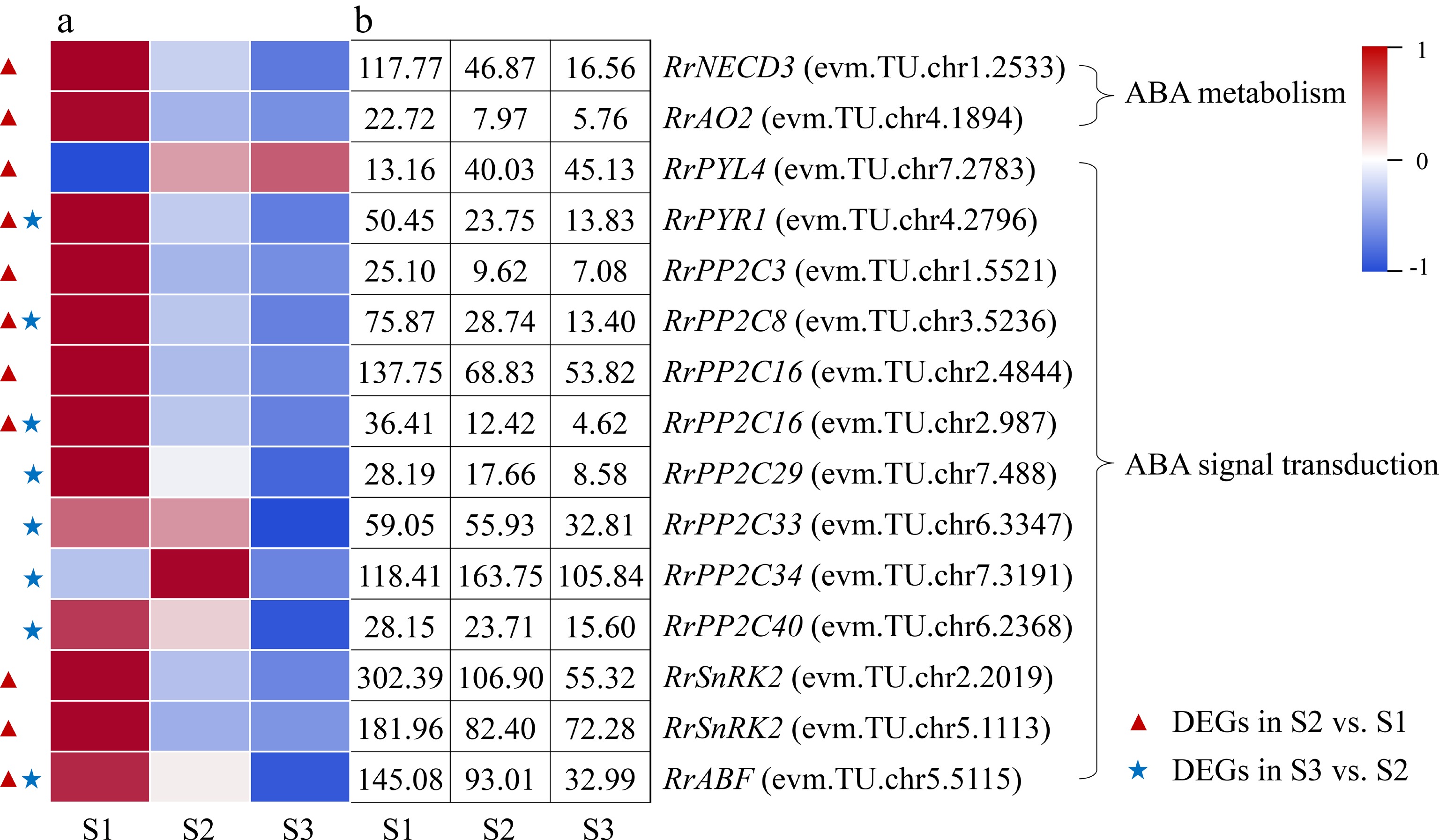
Figure 6.
Expression of ABA-related DEGs. (a) The expression heat map of DEGs. (b) The FPKM values of DEGs. ABF: ABA responsive element binding factor; AO2: Aldehyde oxidase 2; NECD3: 9-cis-epoxycarotenoid dioxygenase 3; PP2C: Protein phosphatase 2C; PYL4: Pyrabactin resistance-like 4 protein; PYR1: Pyrabactin resistance 1 protein; SnRK2: Sucrose nonfermenting 1-related protein kinase 2
-
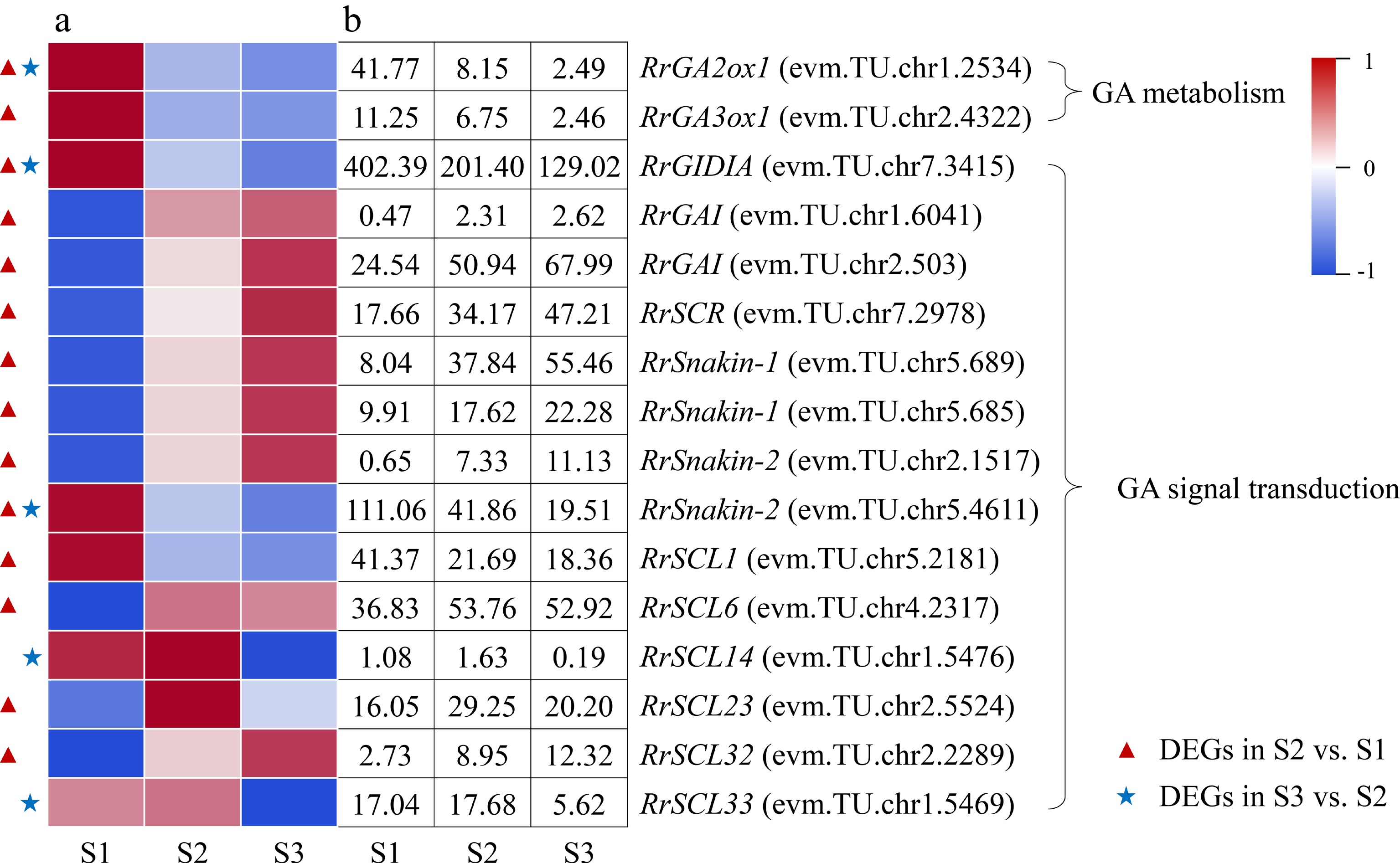
Figure 7.
Expression of GA-related DEGs. (a) The expression heat map of DEGs. (b) The FPKM values of DEGs. GA2ox1: Gibberellin 2-beta-dioxygenase 1; GA3ox1: Gibberellin 3-oxidase 1; GAI: GA insensitive protein; GID1A: GA insensitive dwarf 1 protein; SCL: Scarecrow-like protein; SCR: Scarecrow protein
-
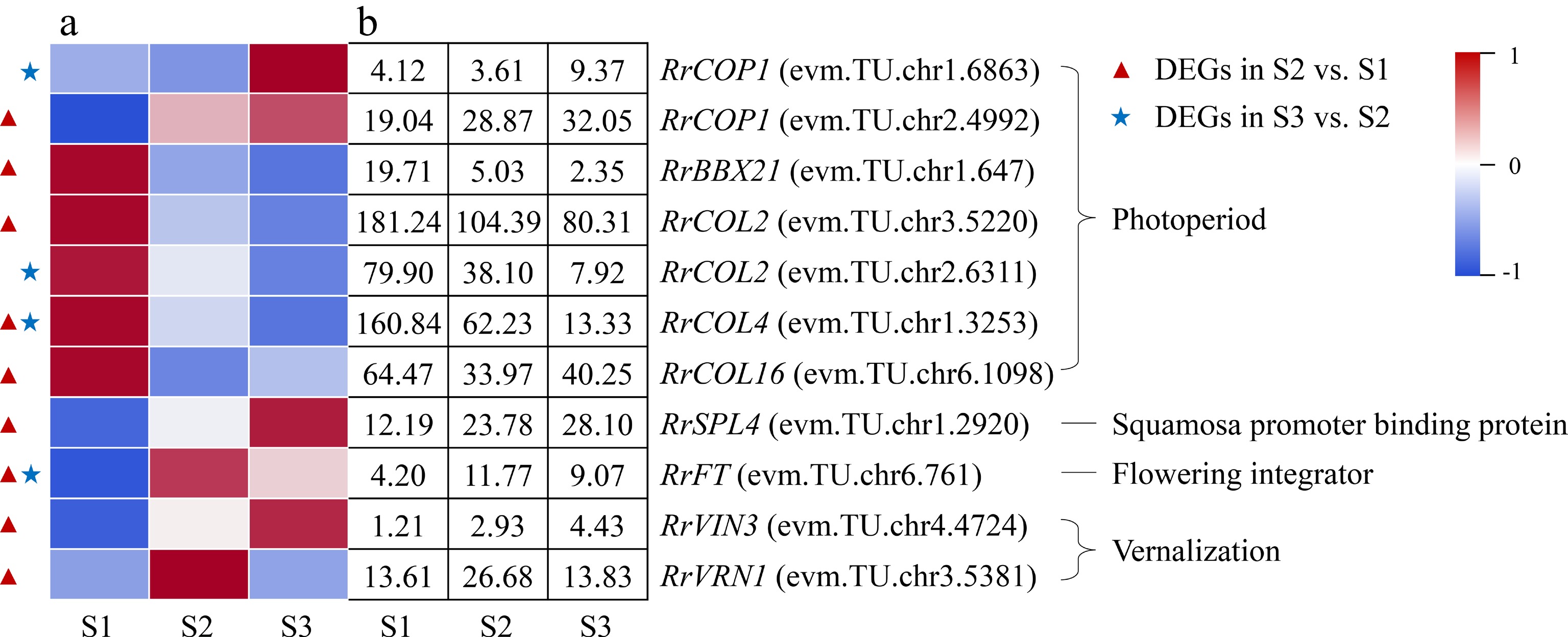
Figure 8.
Expression of photoperiod- and vernalization-related DEGs. (a) The expression heat map of DEGs. (b) The FPKM values of DEGs. BBX21: B-box 21; CO: CONSTANS; COL: CONSTANS-like; COP1: Constitutive Photomorphogenesis 1; FLC: Flowering Locus C; FT: Flowering locus T protein; SPL4: Squamosa promoter-binding-like protein 4;VIN3: Vernalization insensitive protein 3; VRN1: Vernalization protein 1
-
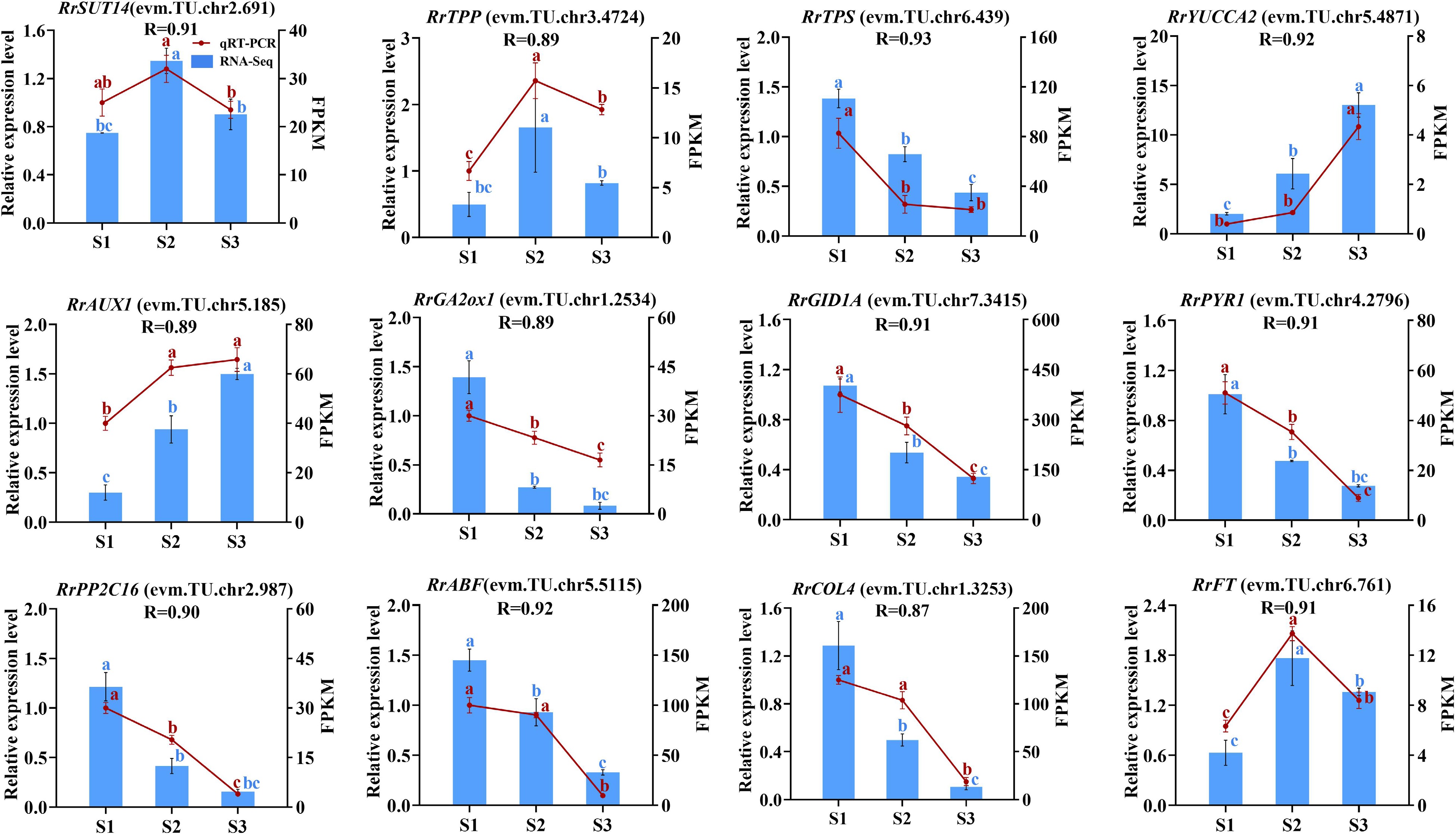
Figure 9.
The correlation of 12 genes between RNA-seq and qRT-PCR data. Letters on the error line indicate significant differences. The different lower case letters for each expression level indicated significant difference between germplasms at ɑ = 0.05.
-
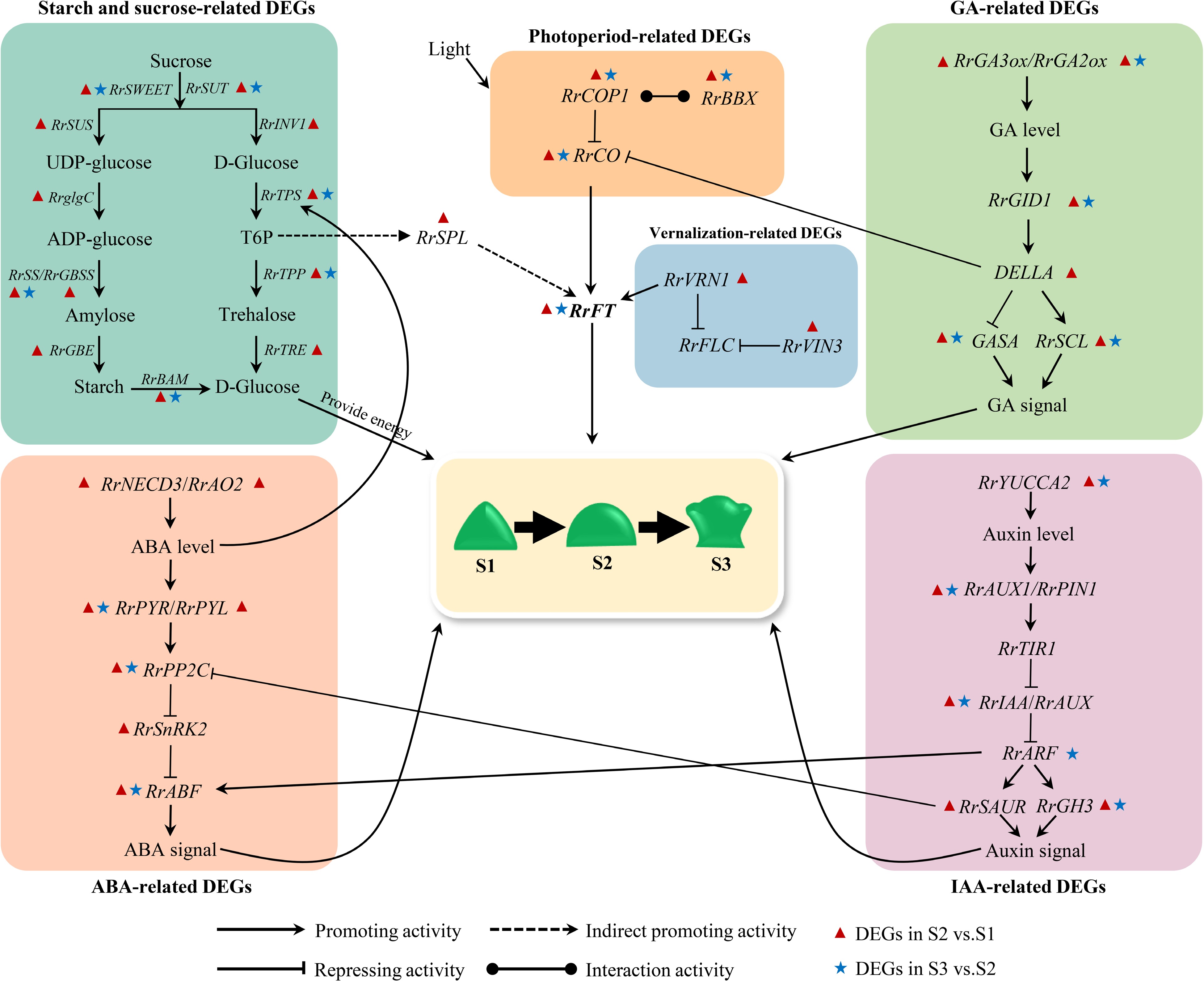
Figure 10.
Hypothetical model for the gene regulatory network of flowering transition in R. rugosa.
Figures
(10)
Tables
(0)