-
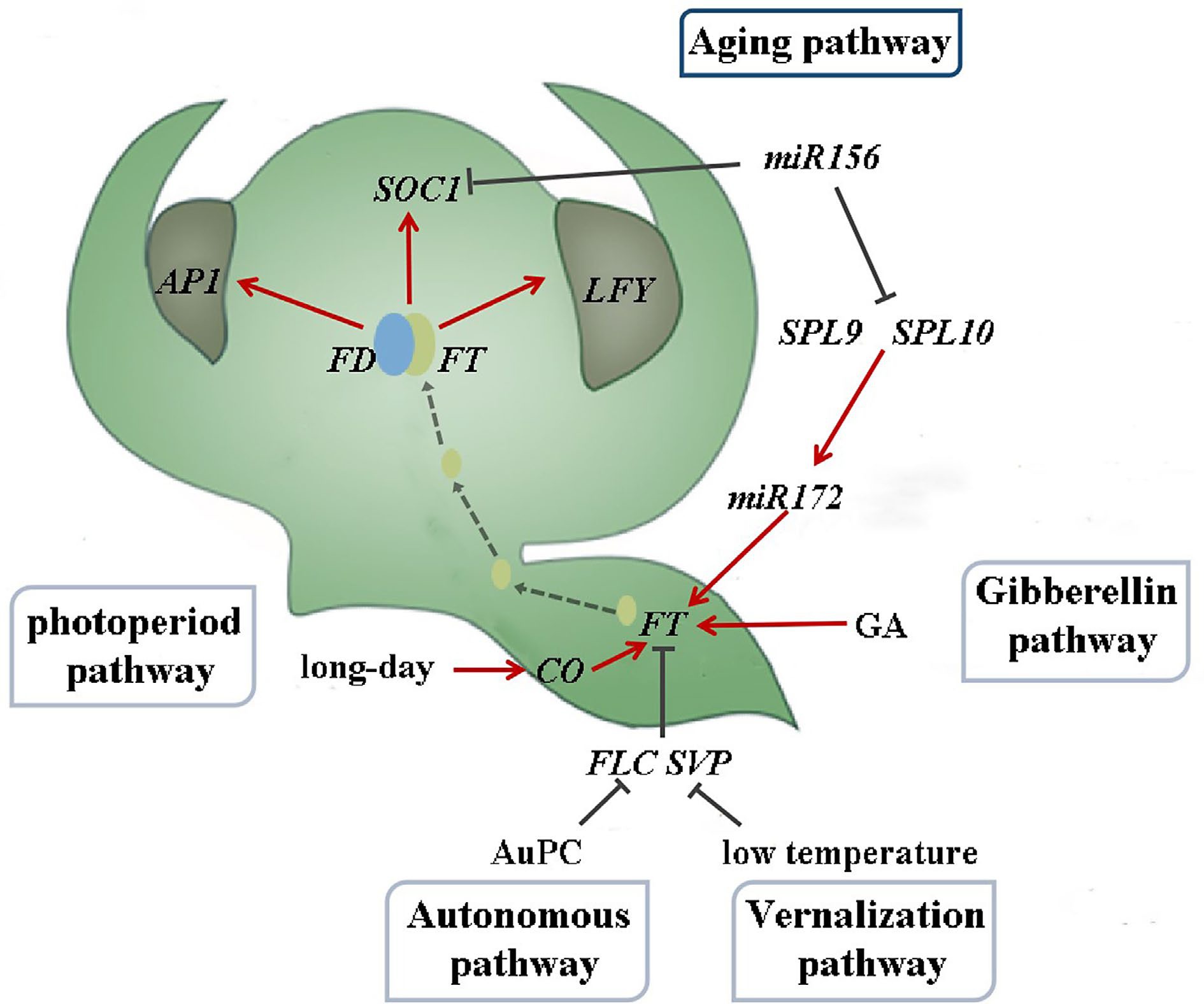
Figure 1.
Flowering induction pathway in Arabidopsis thaliana (drawing from reference[12]).
-

Figure 2.
Vegetative phase change mediated by miR156-miR172 in Arabidopsis thaliana (drawn from reference[12]).
-

Figure 3.
STTM-miR156af interference expression transgenic plants.
-

Figure 4.
MIR172h overexpression transgenic plants.
-
miRNA Target genes Species References miR156 SPL3,4,5,9,10 Arabidopsis thaliana [12, 17,
21, 22]EglSPL3,9 Eucalyptus globulus [43] PcSPL3,9 Populus canadensis PtSPL23,24 Populus tremula × alba [45] MnSPL2,8,10A,10B, 15,16A Morus alba [46] JcSPL2,3,4,5,6,9,10, 11,13,16 Jatropha curcas [47] CiSPL5 Citrus reticulata [48] CmSPL2,4,5,6,9,10,11,13,16,17 Castanea mollissima [49] PgSPL1,2,3,6,7,11,12, 13,14,15 Punica granatum [50] Unigene2030,2872 Cunninghamia lanceolata [51] PeuSPL4,9 Populus euphratica [52] EglSPL3 Eucalyptus globulus [53] PaSPL4,9a,9b Persea americana [54] MiSPL3,4,5 Mangifera indica MciSPL4 Macadamia integrifolia PtSPL1,3 Pinus tabuliformis [56] PaSPL1,2,10,11 Picea abies [57] miR172 AP2;SNZ;SMZ;TOE1,2,3 Arabidopsis thaliana [33, 35] PaAP2,2.7a,2.7b Butyrospermum parkii [54] MiAP2,2.7a,2.7b Mangifera indica MciAP2 Macadamia integrifolia JcAP2;JcTOE1,2,3 Jatropha curcas [58] MsAP2; MsTOE3 Magnolia × soulangeana ‘Changchun’ [59] PtAP2L2,3 Pinus tabuliformis [56] PaAP2L1,3 Picea abies [61] miR159 GAMYB Arabidopsis thaliana [65] MdMYB33,65 Malus pumila [67] PtrMYB012 Populus alba × P. tremula [68] GAMYB Taxus chinensis [69] miR169 NFYA Arabidopsis thaliana [71] PtrHAP2–5 Populus tremuloides [74] Table 1.
miRNAs that regulate flowering time in trees and their target genes.
Figures
(4)
Tables
(1)