-
Plants undergo various developmental transitions throughout their life cycle[1], with the transition from the juvenile to adult phase being particularly significant for flowering plants, where the process of flowering serves as a crucial indicator[2]. Flower development is vital for plant growth and reproduction. The regulatory mechanism of plant flowering is intricate and involves a complex network. The photoperiod, vernalization, autonomous, gibberellin, and aging pathways are the five established pathways for flowering induction in model plants such as Arabidopsis thaliana (Fig. 1)[3−7]. Each pathway regulates gene expression through distinct molecular mechanisms that can interact with one another, culminating in a vast and intricate regulatory network.
Plants undergo a vegetative growth phase until they attain a specific age before they can flower. The aging pathway provides an internal developmental signal that prevents premature flowering and promotes flowering during the adult stage without the need for external factors[8]. This pathway interacts with the photoperiodic, gibberellin, and vernalization pathways[9−11], ultimately regulating plant flowering via the flowering integrator.
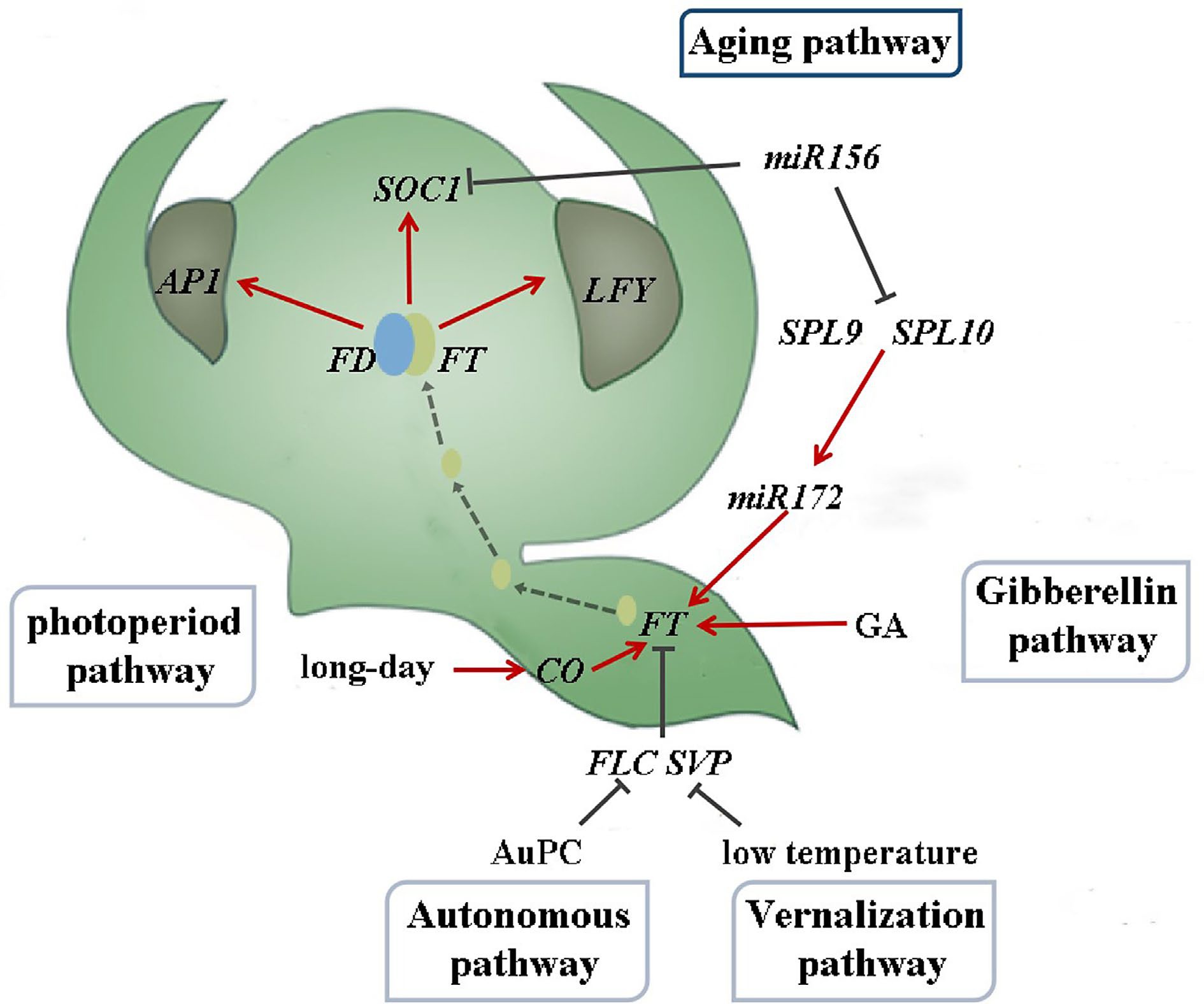
Figure 1.
Flowering induction pathway in Arabidopsis thaliana (drawing from reference[12]).
MicroRNAs (miRNAs) are short, non-coding endogenous RNA molecules that are approximately 18−22 nucleotides in length and play a significant role in plant development and gene regulation[13,14]. Through translational repression or transcriptional cleavage of target genes, miRNAs can repress the expression of target genes, thereby regulating various processes of plant growth and development[15,16]. In recent years, the aging pathway has garnered increasing attention for its role in regulating plant flowering, and miRNAs have been found to control the aging pathway-mediated vegetative phase change[17]. miR156 and miR172 are the central regulatory hub of the aging pathway. They were the first molecular markers discovered in Arabidopsis that respond to aging pathway regulation[18] and have been systematically studied in species such as Zea mays and Chrysanthemum morifolium, providing an important approach to explore and reveal the regulatory mechanisms of plant growth and development.
Currently, the miRNA156 and miRNA172-mediated aging pathway has been extensively studied in model plants, but there is limited research on perennial trees. Perennial trees are characterized by a long juvenile phase and delayed flowering and fruiting, which significantly impacts their reproductive efficiency and limits the breeding process. Therefore, it is necessary to investigate the age-regulated pathways in trees. This review primarily focuses on the role of miRNA156 and miRNA172 in vegetative phase transition in trees and discusses the involvement of other miRNAs outside the aging pathway. This review also covers the flowering-related miRNAs and their target genes, as well as the mode of action between miRNAs.
-
miR156 is a highly conserved and widely studied member of the plant miRNA family. It was first discovered in Arabidopsis[19] and is involved in regulating plant phenotypic changes and flowering through its interaction with the SPL family[19,20]. The 3' UTR of SPL family members contains a functional miRNA response element (MRE), which can be targeted by miR156 to promote degradation or repression of translation of target genes[21].
Expression of miR156 is high during the seedling stage and gradually decreases as the plant develops[22]. In Arabidopsis, the miR156 family consists of eight members, with miR156a and miR156c playing a dominant role in vegetative phase change[23]. The importance of miR156 in flowering is demonstrated by the delayed flowering phenotype resulting from miR156 overexpression[24]. Loss of function of miR156 in STTM (short tandem target mimic) lines in Arabidopsis leads to an early flowering phenotype, with upregulation of miR172, SPL, and some flowering activators in the STTM-miR156 lines[25].
A total of 17 SPL genes were identified in Arabidopsis, 11 of which are direct downstream target genes of miR156[21]. Currently, the miR156-SPL module was found to affect the flowering process in plants mainly through the regulation of downstream related flowering genes. There are two main pathways through which miR156 regulates flowering time in Arabidopsis: first, in SAM (shoot apical meristem), the target genes of miR156, SPL3 and SPL9, induce flowering by activating floral organ characteristic genes such as SOC1 and AP1. The other pathway is in leaves, where SPL9 activates the expression of miR172 genes and further suppresses the expression of flowering repressors such as AP2 to accelerate flowering[12].
Molecular mechanism of miR172
-
miR172, a downstream target of miR156, plays a crucial role in the transition to the adult phase in plants[26]. This miRNA is highly conserved in ferns, gymnosperms, and angiosperms[27]. Multiple studies have shown that miR172 regulates flowering time, floral organ development, and plays a crucial role in vegetative phase change in several species[28]. In Arabidopsis, the precursor of miR172 is encoded by miR172a-miR172e. Overexpression of miR172 in Arabidopsis has been found to promote the transition from the juvenile to adult phase[29−31].
The expression of plant miR172 increases during growth and development, while the expression of its target genes decreases, indicating that miR172 primarily functions by repressing translation of its targets[32]. Its targets include AP2/ERF transcription factors[33], with six target genes identified in Arabidopsis: AP2, SNZ, SMZ, TOE1, TOE2, and TOE3[34]. Through interaction with the conserved structural domains of these target genes, miR172 promotes flower development by suppressing their accumulation[35].
In model plants such as Arabidopsis, the miR156-SPL-miR172-AP2 pathway is well understood (Fig. 2). The miR156 gene family indirectly regulates miR172 expression by suppressing SPL gene expression, which in turn reduces the transcription of downstream genes like AP2 and TOE. This regulatory pathway plays a crucial role in all plant development processes[36].

Figure 2.
Vegetative phase change mediated by miR156-miR172 in Arabidopsis thaliana (drawn from reference[12]).
-
Plants exhibit distinct morphological features during various developmental stages. For instance, Arabidopsis shows differences in leaf morphology between juvenile and adult stages, with adult leaves having serrations and abaxial trichomes[37]. In contrast, perennial woody plants with longer developmental cycles display subtle differences between their juvenile and adult stages. In such plants, the vegetative phase change has been studied by examining phenotypic characteristics such as leaf morphology, stem morphology, and branching. Leaf morphology was first investigated as early as 1990, revealing that plants displayed varying phenotypic features and sizes, photosynthetic efficiency, or epidermal characteristics at different developmental stages[38]. Recent studies have also examined the vegetative phase change in poplar[39] and apple[40] species, and investigated the developmental status of trees at the stem[41] and branches[42].
In some heteroblastic species, including Eucalyptus globulus, Acacia confusa, Quercus acutissima, and Acacia colei, the transition from juvenile to adult phases is marked by distinct morphological changes. For instance, Acacia trees shift from pinnate compound leaves to produce simple undivided leaves known as phyllodes. The expression patterns of miR156 in juvenile and adult leaves of these species were analyzed separately, and it was found that miR156 expression decreased with age, consistent with the pattern observed in Arabidopsis. Additionally, miR156 expression levels correlated with changes in leaf morphology and internode distance, indicating that miR156 is associated with the vegetative phase change. Overexpression of miR156 in Populus canadensis suppressed the expression of SPL and miR172 in transgenic lines and significantly prolonged the juvenile period, further supporting the role of miR156 in phase change[43]. Similarly, in Populus tremula × P. alba, overexpression of Cg1, a maize endogenous gene encoding miR156, resulted in significant branching and reduced lignin content in transgenic lines. Furthermore, a positive correlation was observed between the severity of the phenotypic changes and Cg1 expression levels, underscoring the significant effects of miR156 on poplar growth and development[44]. A study investigating miR156-SPL expression regulation patterns in Populus tremula × P. alba found that lower miR156 expression levels in leaves of different node positions were associated with smaller leaf length-width ratios, while transgenic lines with high miR156 expression levels had smaller leaf areas and shorter petiole lengths, further confirming the role of miR156 in the vegetative phase change of poplar[45].
Research on various plant species including Morus alba, Jatropha curcas, Citrus reticulata, Castanea mollissima, Punica granatum, and Cunninghamia lanceolata has confirmed the existence of the miR156-SPL regulatory network which plays an important role in plant vegetative phase change. In mulberry, miR156 expression gradually decreases with age while SPL and miR172 expression show the opposite trend. Studies have shown that the miR156-SPL-miR172 regulatory network is present in mulberry and can activate the expression of miR172a by recognizing the GTAC cis element in the promoter region[46]. In Jatropha curcas, JcSPL3 plays an important role in regulating age development, with its expression level increasing with plant age. Overexpression of JcSPL3 in Arabidopsis results in earlier flowering, validating its function[47]. Similarly, in Citrus reticulata, CiSPL5 plays a major role in promoting flowering and accelerating the juvenile process, regulated by miR156[48]. Genome-wide identification of the SPL gene family in Castanea mollissima[49], Punica granatum[50], and Cunninghamia lanceolata[51] further supports the importance of the miR156-SPL regulatory model in plant vegetative phase change.
As Populus euphratica leaves matured, they transitioned from a lanceolate to a broad-ovate shape, and this process was associated with a decrease in miR156 expression and an increase in the expression of 12 SPLs, suggesting that the miR156-SPL regulatory pathway plays an important role in the plant's vegetative phase change[52]. Similarly, in Eucalyptus globulus, differences in miR156 expression were found to underlie the timing of the plant's vegetative phase change, as indicated by QTL analysis of an outcross F2 family[53]. In studies of Persea americana, Mangifera indica, and Macadamia integrifolia, miR156 expression gradually decreased with tree age, and the miR156-SPL3/4/5 pattern was found to be conserved in all three species, consistent with previous studies[54].
The aging pathway has been investigated in gymnosperms. In Ginkgo biloba, miRNA expression changes at different ages were analyzed, revealing that miR156 was highly expressed in older individuals, and its target genes did not vary with age, unlike the miR156 expression pattern observed in angiosperms[55]. In Pinus tabuliformis, 26 miRNA families and 74 targets were identified through small RNA sequencing and parallel analysis of RNA ends (PARE). Among these miRNAs, miR156-SPL, which is involved in angiosperm reproductive development, was also found to be present in P. tabuliformis[56]. Picea abies genome analysis identified 11 SPL genes, PaSPL1-11, among which PaSPL1, PaSPL2, PaSPL10, and PaSPL11 have miR156 binding sites. The evolutionary relationships suggested that PaSPL1, PaSPL10, and PaSPL11 play a crucial role in reproductive stage change, but miR156-SPL expression patterns vary in different tissues. Additionally, mutations in the miR156 binding site of the PaSPL1 gene were found to cause early flowering of the acrocona mutant, demonstrating the importance of the aging pathway in regulating reproductive phase change in P. abies[57]. These studies show that miR156 and its target gene SPLs are conserved in gymnosperms, but no age-related expression pattern of miR156-SPL was observed in gymnosperms, suggesting that there is no trend of decreasing or increasing expression levels of miR156 or SPL with increasing plant age in gymnosperms.
Our team studied the precursor and mature sequences of the miR156 gene family using the Populus tomentosa genome data, and we were able to clone 11 members of the family, which we designated pto-miR156a/b/c/d/e/f/g/h/i/j/k based on comparison results. After that, we analyzed the family members' sequences, predicted secondary structure and target genes, and examined their expression patterns. We created an expression interference vector for STTM-miR156af based on the mature sequence's conserved domain. We identified DNA and RNA levels in transgenic lines produced by genetic transformation mediated by Agrobacterium. Our study revealed that the transgenic lines exhibited a significant reduction in the transcription of miR156a/b/c/d/e/f when compared to the wild type. Additionally, we found that the plant height of the transgenic lines was 35.44% shorter than that of the wild type (Fig. 3), indicating that miR156 plays a crucial role in plant height regulation. These findings are in line with previous studies that utilized STTM technology to repress miR156a in poplar[45] and observed a decrease in plant height in the transgenic line relative to the wild type.
To gain further insights into the regulatory mechanism of miR156 in the aging pathway of poplar, we plan to conduct statistical analysis of related phenotypes in leaves and other aspects. Furthermore, we intend to integrate sequencing data to explore the complex interplay between miR156 and aging-related genes in poplar. Our study will provide a comprehensive understanding of the role of miR156 in regulating plant growth and aging and offer potential targets for tree improvement.
The role of miR172 and its target gene AP2-like in trees
-
Changes in miR172 were observed in the leaves of various plants such as E. globulus, A. confusa, Q. acutissima, and A. colei[43]. Unlike miR156, miR172's expression increased with age and was linked to leaf morphology and internode distance, indicating its role in regulating the vegetative phase change. Overexpression of miR156 in P. canadensis suppressed the expression of miR172, prolonging its juvenile phase. Conversely, miR172's expression gradually increased with age in poplar, indicating its role in regulating the vegetative phase change[43]. In mulberry, miR172's expression was found to be proportional to the tree's age, and six miR156 target genes activated miR172 expression[46]. The study of miR172 in J. curcas revealed that its expression increased with tree age, and its overexpression caused early flower and leaf morphological changes, while suppressing the expression of miR172 target genes[58]. In Magnolia × soulangeana ‘Changchun’, miR172 expression levels were significantly upregulated during the transition from vegetative to reproductive growth, and its target genes MsAP2 and MsTOE3 were downregulated, indicating its significant role in the vegetative phase change of magnolia through molecular regulation[59].
In more mature tissues of avocado (Butyrospermum parkii), miR156 was found to be more abundant, while miR172 was less abundant compared to young tissues. The expression of their target genes, PaSPL4 and PARA2.7B, respectively, was found to be negatively correlated with the abundance of miR156 and miR172. These findings suggest the conservation of the miR156-SPL-miR172 pathway in avocado[60]. However, a different study showed that although miR172 abundance significantly increased during the transition from juvenile to adult avocado, the expression of AP2-like genes did not decrease with increasing miR172 abundance. As this study was based on RNA transcript abundance, it cannot conclusively rule out the possibility of a translational repressive effect of miR172[54].
The expression of miR172 and its target gene AP2 was investigated in different age groups of Ginkgo biloba, a gymnosperm. The study showed that unlike in angiosperms, miR172 and its target genes did not exhibit an age-related differential expression pattern[55]. In Pinus tabuliformis, miR172-AP2 was also identified through small RNA sequencing and parallel analysis of RNA ends (PARE) in both female and male cones[56]. Homologs of AP2 in Arabidopsis, PaAP2L1, PaAP2L2, and PaAP2L3, were identified in Picea abies, and it was confirmed that all three genes contained sequence motifs complementary to miR172. The overexpression of PaAP2L2 in Arabidopsis resulted in delayed development in transgenic lines, supporting the conservation of AP2 in Picea abies[61]. In conclusion, miR172-AP2 is conserved in gymnosperms, but unlike in angiosperms, no clear expression pattern was observed.
Using genome data from P. tomentosa, our group analyzed the precursor and mature sequences of the miR172 gene family. We identified seven members of the miR172 family, which we named Pto-miR172a/b/c/e/g/h/i based on comparative analysis. We conducted sequence and expression pattern analyses of these seven members and selected key members to construct a C6-MIR172h overexpression vector. We then carried out genetic transformation in poplar and observed a 26.82% increase in height growth in the transgenic line compared to the wild type (Fig. 4). These results are consistent with previous findings on the role of miR172 in regulating plant height in cotton[62] and rice[63]. We will further analyze the phenotypic data of the transgenic lines and explore the mechanisms of miR172 in combination with transcriptomic data. This will provide a basis for elucidating the aging pathway of poplar flowering regulation.
-
In addition to miR156 and miR172, other miRNAs involved in regulating flowering have been reported in recent years, including miR159 and miR169. While these miRNAs are not part of the aging pathway, they interact with miR156/172 to jointly regulate plant flowering.
The miR159-Regulated Pathway
-
miR159 is a highly conserved microRNA family that is commonly found in plants[64]. Studies have shown that miR159 targets a class of GAMYB genes that encode R2R3-MYB transcription factors. These GAMYB genes activate downstream genes related to GA signaling and are also referred to as 'GAMYB' genes[65]. In Arabidopsis thaliana, miR159 regulates the timing of the vegetative phase change by suppressing the expression of MYB33, preventing it from hyperactivating miR156 expression during the seedling stage[66].
The mechanism of miR159 action has also been explored in trees in recent years. The expression of miR159a increased significantly during the dormancy transition in Malus pumila, whereas the expression of its putative target genes MdMYB33 and MdMYB65 decreased substantially during this period, which also suggests that miR159-MYB plays an important role during dormancy in M. pumila, regulating M. pumila break dormancy to further promote floral bud differentiation[67]. Although PtrMYB012 contains a conserved miR159 target sequence, it is not a target of Arabidopsis miR159. Therefore, its transcript is not degraded and overexpression of PtrMYB012 in Arabidopsis leads to phenotypes such as upwardly curled rosette leaves and plant dwarfism. In contrast, PtrMYB012 was affected by poplar miR159, resulting in degradation of transcripts, and ultimately no significant phenotypes occurred in transgenic poplar, suggesting that miR159-MYB in poplar regulates plant development during the vegetative phase[68]. miR159 is one of the most abundantly expressed miRNAs during different developmental periods in Taxus chinensis, and studies have demonstrated that it effectively represses GAMYB and thus regulates the growth and development of T. chinensis[69].
The miR169-regulated pathway
-
Currently, miR169, which is the largest miRNA gene family in plants, has been extensively studied in relation to plant growth and development, resistance, and regulation of flowering[70−72]. The NFYA family is the most important target of miR169. The miR169-NFYA module also plays a crucial role in the development of floral organs in plants. In Arabidopsis, AtNFYA2 binds to and activates the expression of the AACCT component of the flowering repressor FLC (flowering locus C) promoter, thus delaying the flowering time of plants[71]. Similarly, the binding of Arabidopsis NFYA8 to the miR156 promoter prevents the transition of plant seedlings to adults and delays the flowering time[73].
The role of miR169 in regulating flowering has been investigated in Populus tremuloides. The study found a negative correlation between the expression dynamics of ptr-miR169a and its target gene PtrHAP2-5, particularly at the beginning of dormancy when ptr-miR169a expression increased and PtrHAP2-5 transcript levels decreased. The study also verified that the transcriptional decline of PtrHAP2-5 was caused by cleavage at the predicted miR169a site. It was suggested that HAP2 has an opposite effect on FT transcriptional activity. Moreover, miR169 was found to alleviate the repression of the PtrFT gene during dormancy by down-regulating the abundance of PtrHAP2 transcripts, thus promoting plant flowering[74].
-
This review critically examines recent research on miRNAs and their roles in regulating the flowering of trees, with a particular focus on miR156 and miR172 in the aging pathway, along with miR159 and miR169, which interact with the aging pathway to promote flowering (as outlined in Table 1). Based on the analysis of the findings, it was observed that miR156 and miR172 are commonly present in angiosperms, and their target genes exhibit either an increase or decrease in expression as the plant ages. However, there is no evidence to support age-related expression patterns for these miRNAs in angiosperms.
Table 1. miRNAs that regulate flowering time in trees and their target genes.
miRNA Target genes Species References miR156 SPL3,4,5,9,10 Arabidopsis thaliana [12, 17,
21, 22]EglSPL3,9 Eucalyptus globulus [43] PcSPL3,9 Populus canadensis PtSPL23,24 Populus tremula × alba [45] MnSPL2,8,10A,10B, 15,16A Morus alba [46] JcSPL2,3,4,5,6,9,10, 11,13,16 Jatropha curcas [47] CiSPL5 Citrus reticulata [48] CmSPL2,4,5,6,9,10,11,13,16,17 Castanea mollissima [49] PgSPL1,2,3,6,7,11,12, 13,14,15 Punica granatum [50] Unigene2030,2872 Cunninghamia lanceolata [51] PeuSPL4,9 Populus euphratica [52] EglSPL3 Eucalyptus globulus [53] PaSPL4,9a,9b Persea americana [54] MiSPL3,4,5 Mangifera indica MciSPL4 Macadamia integrifolia PtSPL1,3 Pinus tabuliformis [56] PaSPL1,2,10,11 Picea abies [57] miR172 AP2;SNZ;SMZ;TOE1,2,3 Arabidopsis thaliana [33, 35] PaAP2,2.7a,2.7b Butyrospermum parkii [54] MiAP2,2.7a,2.7b Mangifera indica MciAP2 Macadamia integrifolia JcAP2;JcTOE1,2,3 Jatropha curcas [58] MsAP2; MsTOE3 Magnolia × soulangeana ‘Changchun’ [59] PtAP2L2,3 Pinus tabuliformis [56] PaAP2L1,3 Picea abies [61] miR159 GAMYB Arabidopsis thaliana [65] MdMYB33,65 Malus pumila [67] PtrMYB012 Populus alba × P. tremula [68] GAMYB Taxus chinensis [69] miR169 NFYA Arabidopsis thaliana [71] PtrHAP2–5 Populus tremuloides [74] Currently, our understanding of the regulatory mechanisms of flowering in plants has largely been acquired from annual model plants. However, the flowering process in perennial trees is more complex due to seasonality and annual cycles, which may involve additional and different regulatory mechanisms. Unfortunately, research on elucidating flowering mechanisms in perennial plants has progressed slowly due to complications arising from their perennial growth and the recurring nature of the flowering process. Although studies on flowering mechanisms have been carried out in some perennial horticultural plants or crops, these studies have provided genetic resources and cases for flowering studies in perennials, deepening our understanding of flowering mechanisms. Despite this progress, relatively few studies have focused on the molecular mechanisms of flowering regulation in tall tree species, and age-regulated pathways have barely been reported. miRNAs are known to play an important role in regulating flowering as well as overall growth and development in plants. Therefore, understanding the molecular mechanisms of miRNA action in perennial trees holds great research and practical value. It is necessary to deepen our research on the mechanisms of miRNA action in trees to advance the breeding process.
This work was supported by the China National Key R&D Program (2021YFD2200101), Major Project of Agricultural Biological Breeding (2022ZD0401503), and the National Natural Science Foundation of China (31870652, 31570661).
-
The authors declare that they have no conflict of interest.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Li Y, Chen T, Khan WU, An X. 2023. Regulatory roles of miRNAs associated with the aging pathway in tree vegetative phase changes. Forestry Research 3:9 doi: 10.48130/FR-2023-0009
Regulatory roles of miRNAs associated with the aging pathway in tree vegetative phase changes
- Received: 03 December 2022
- Accepted: 17 March 2023
- Published online: 18 April 2023
Abstract: The transition from the vegetative juvenile phase to the adult phase is a crucial event in the life cycle of flowering plants, with flowering being the most important milestone. While the regulatory pathways of flowering have been well established in model plants such as Arabidopsis and a few crops, the flowering regulation pathways in perennial forest trees remain poorly understood. This paper summarizes the regulation of flowering time by miR156 and miR172, which are the main members of the aging pathway, and also presents new information on the role of miR159 and miR169. These two microRNAs interact with miR156 and miR172 to jointly regulate flowering time in forest trees. Overall, this review sheds light on the complex regulatory mechanisms underlying flowering time in forest trees and provides insights into potential targets for manipulating the flowering time of these economically and ecologically important species.
-
Key words:
- Flowering regulation /
- Forest tree /
- miR156 /
- miR172















