-
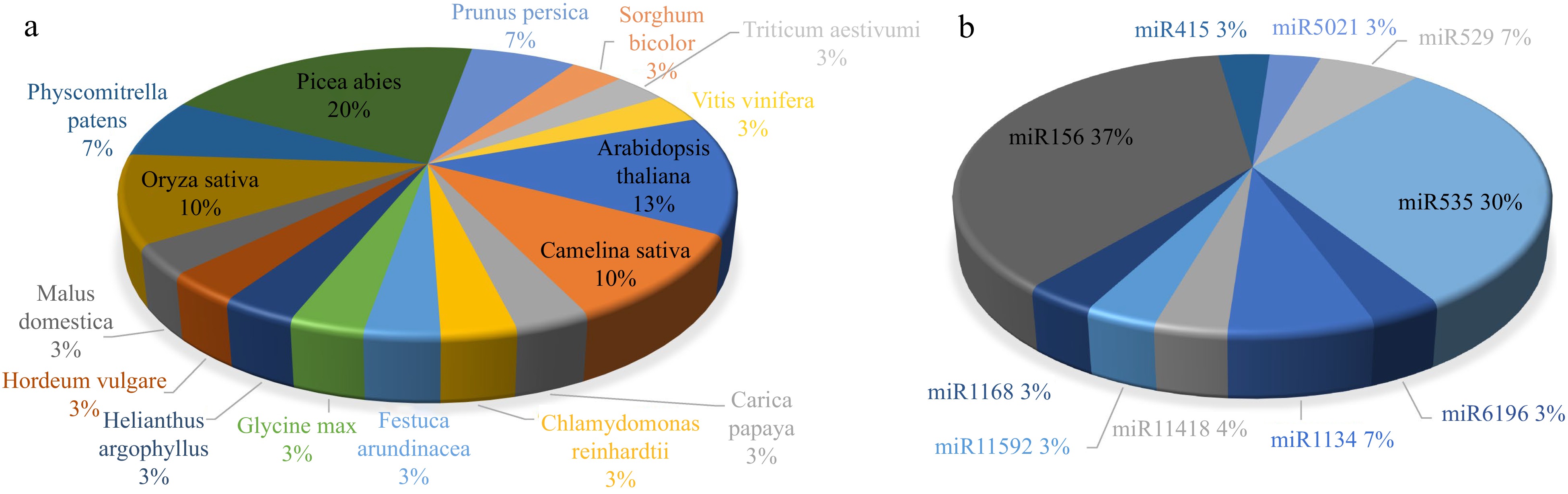
Figure 1.
(a) Potential microRNAs found by bowtie software come from known microRNAs of different species. (b) Potential microRNAs found by bowtie software from known microRNAs families.
-
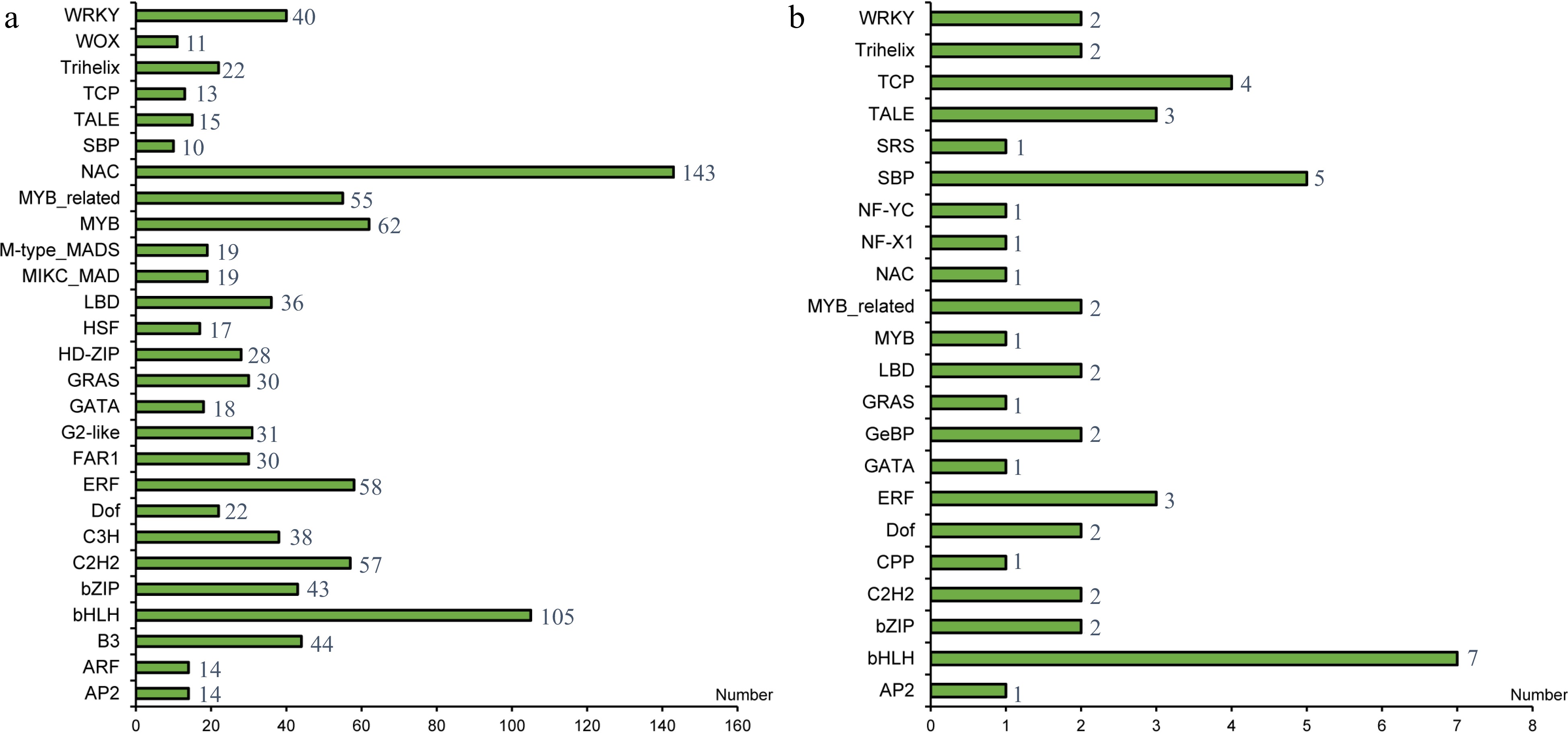
Figure 2.
(a) Number of transcription factor families in S. japonica. (b) Statistical analysis of 47 transcription factors with regulatory relationship with microRNA.
-
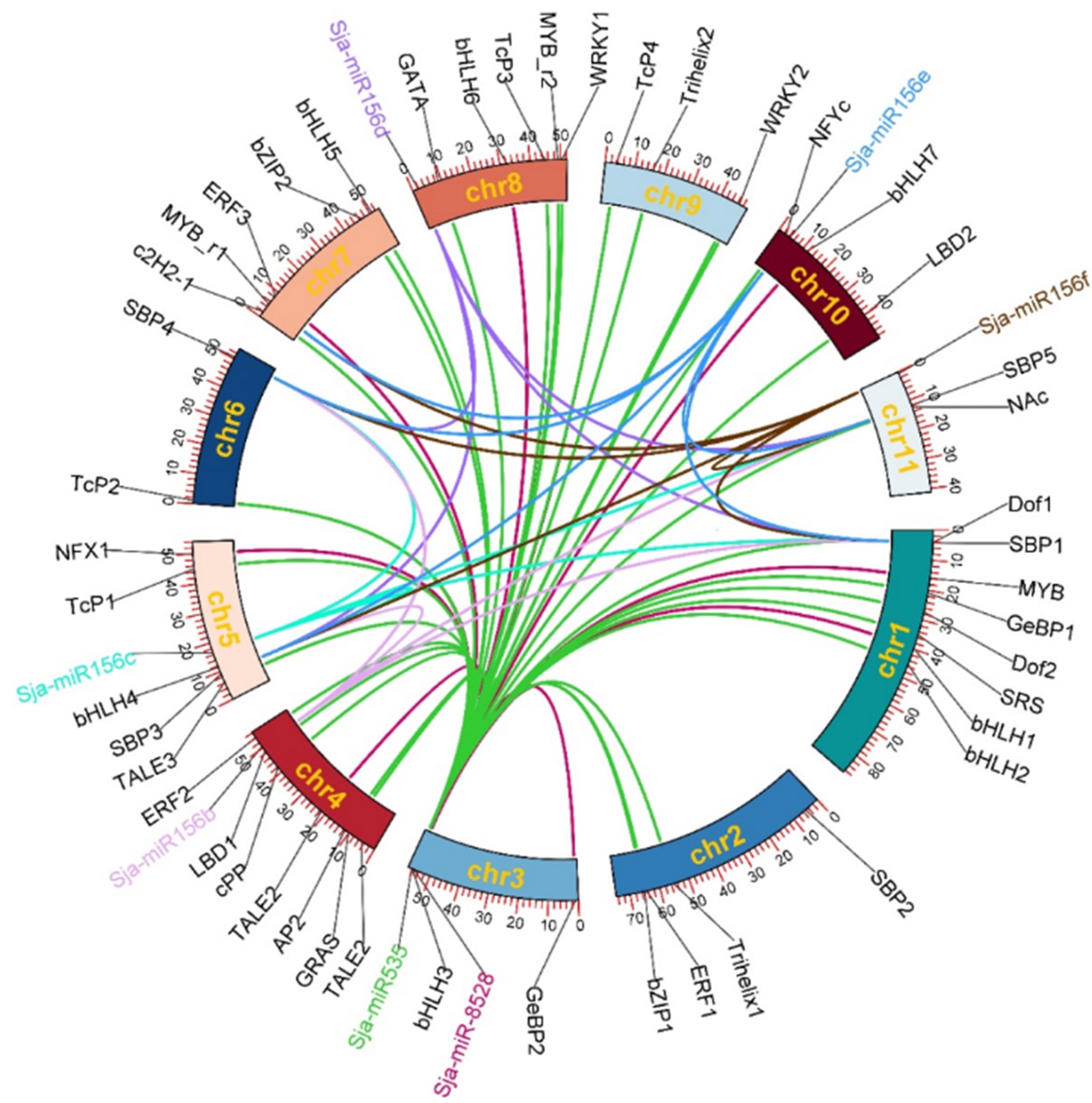
Figure 3.
The chromosome mapping of microRNA and transcription factors.
-
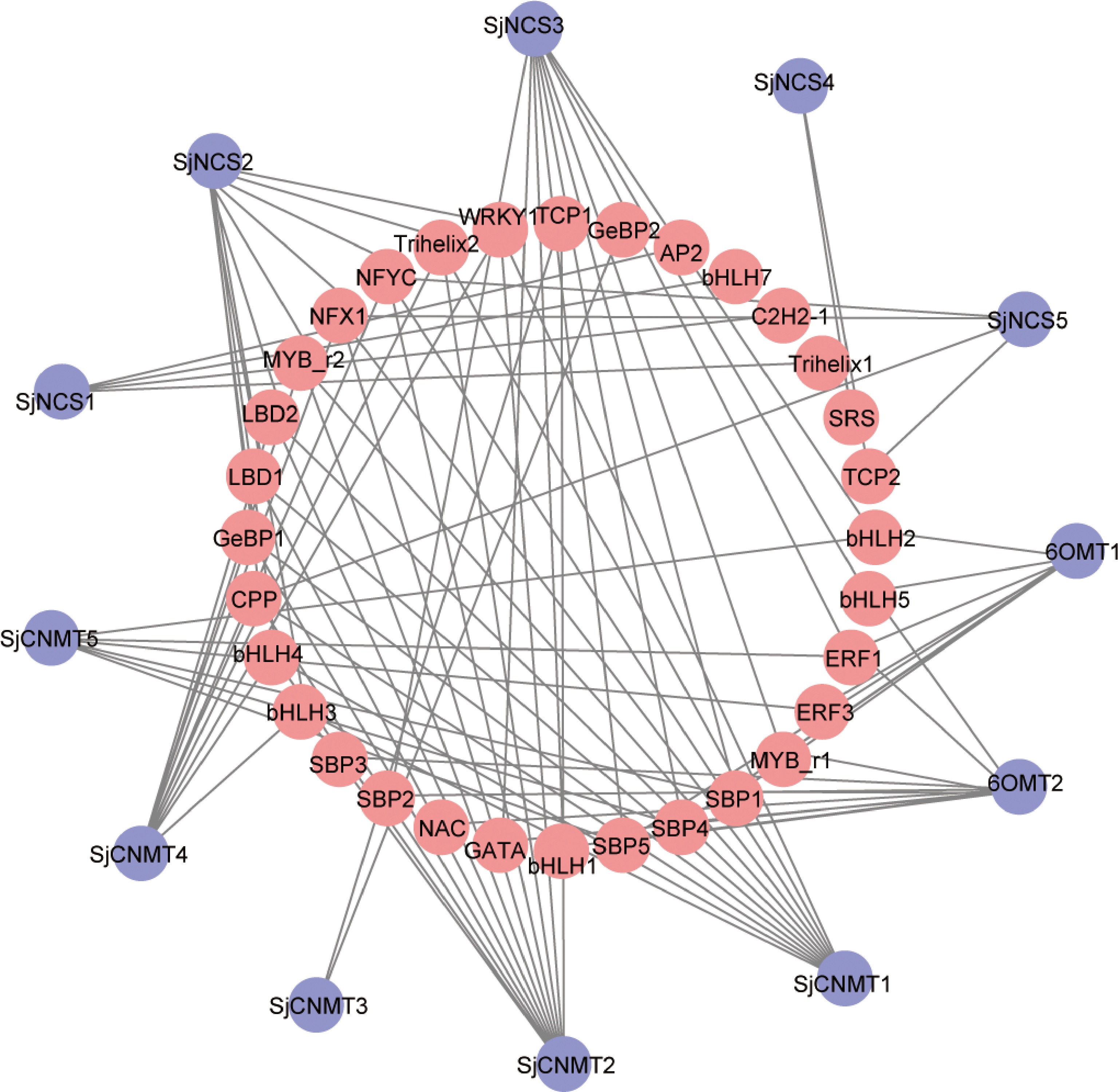
Figure 4.
Regulatory network of transcription factors genes with CEP biosynthetic genes in S. japonica. Red: transcription factors genes. Purple: CEP biosynthetic genes.
-
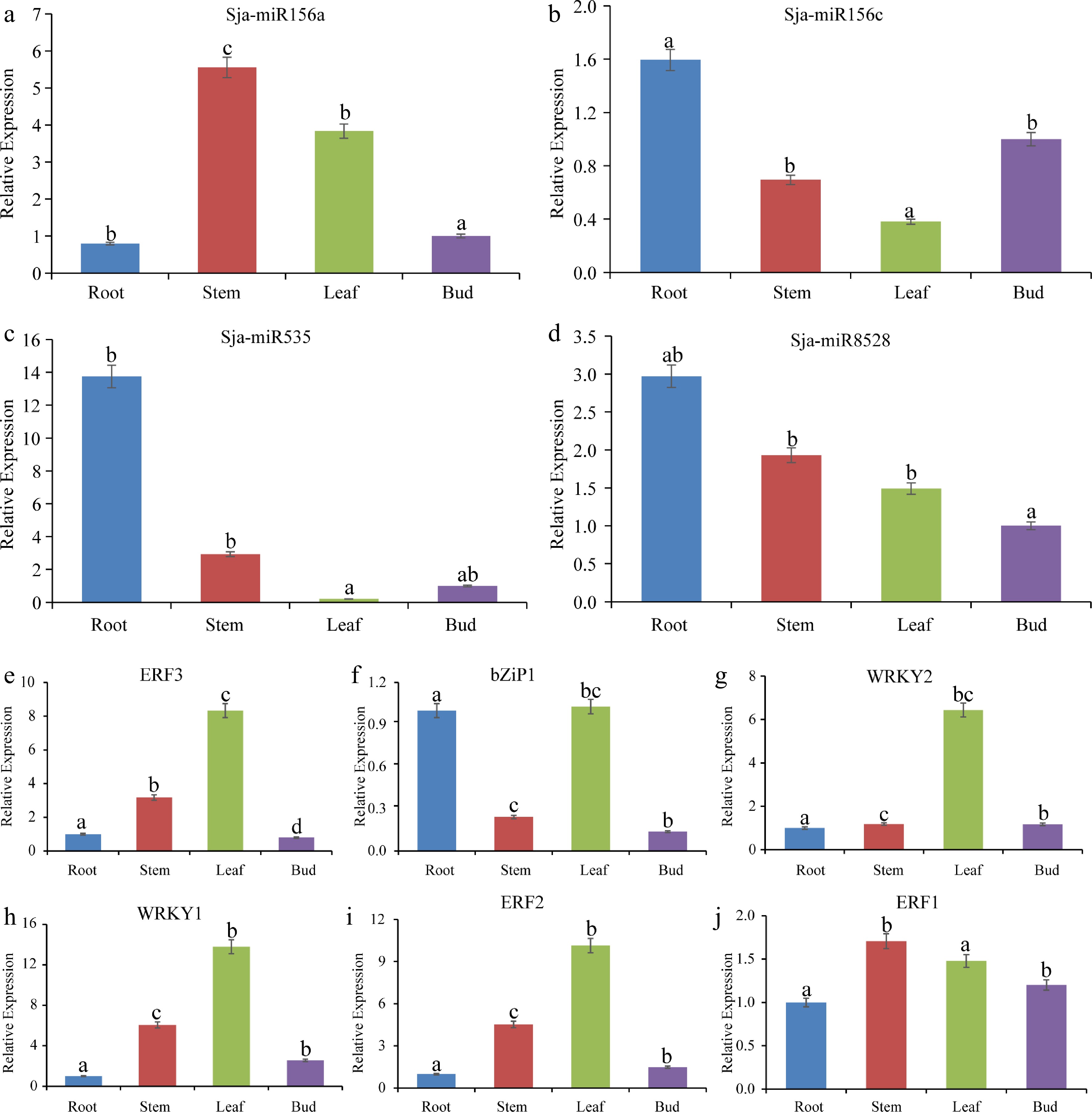
Figure 5.
Expression analysis of microRNA and transcription factors in different tissues by qRT-PCR. (a) - (d) Four microRNA expression levels. (e) - (j) Six transcription factors expression levels. Different lowercase letters indicate significant differences between different treatments (P < 0.05).
-
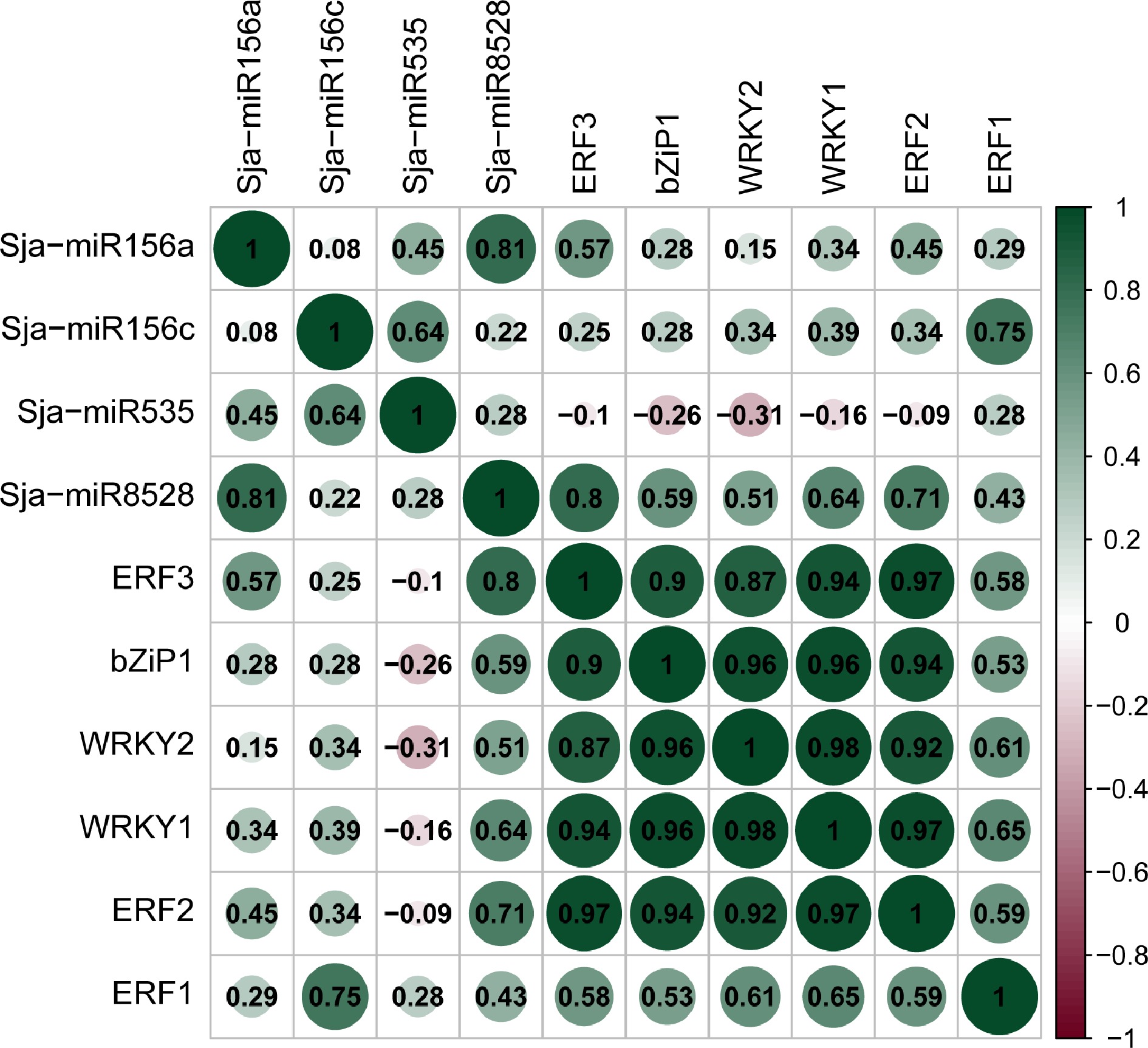
Figure 6.
Correlation analysis of microRNAs and transcription factors.
-
Name LM
(nt)Query
microRNAChromosome
positionmicroRNA
sequencesAccession Loc LP
(nt)GC
(%)MFEs
(ΔG)MFEI Sja-miR156a 20 ath-miR156h chr10_4372847_4373267 UGACAGAAGAAAGAGAGCAC MIMAT0001013 5' 85 44 −45 1.2 Sja-miR156b 20 ath-miR156h chr11_610113_610533 UGACAGAAGAAAGAGAGCAC MIMAT0001013 5' 84 39 −42.2 1.3 Sja-miR156c 19 cas-miR156g chr4_50504862_50505281 GACAGAAGAGAGUGAGCAC MIMAT0045276 5' 81 48 −48.6 1.3 Sa-jmiR156d 19 cas-miR156g chr5_16609851_16610270 GACAGAAGAGAGUGAGCAC MIMAT0045276 5' 81 48 −48.6 1.3 Sja-miR156e 19 cas-miR156g chr8_1964637_1965056 GACAGAAGAGAGUGAGCAC MIMAT0045276 5' 84 50 −36.3 0.9 Sja-miR535 20 ppt-miR535a chr3_56476014_56476435 GGCGGCGGCGAGGCGGGGG MIMAT0003139 5' 81 56 −46.5 1.0 Sja-miR8528 18 pxy-miR-8528a chr3_55723033_55723451 GGGCGGCGGCGAGCGGGA MIMAT0033748 5' 49 80 −48.2 1.2 Table 1.
Summary of the identified microRNAs from S. japonica.
Figures
(6)
Tables
(1)