-
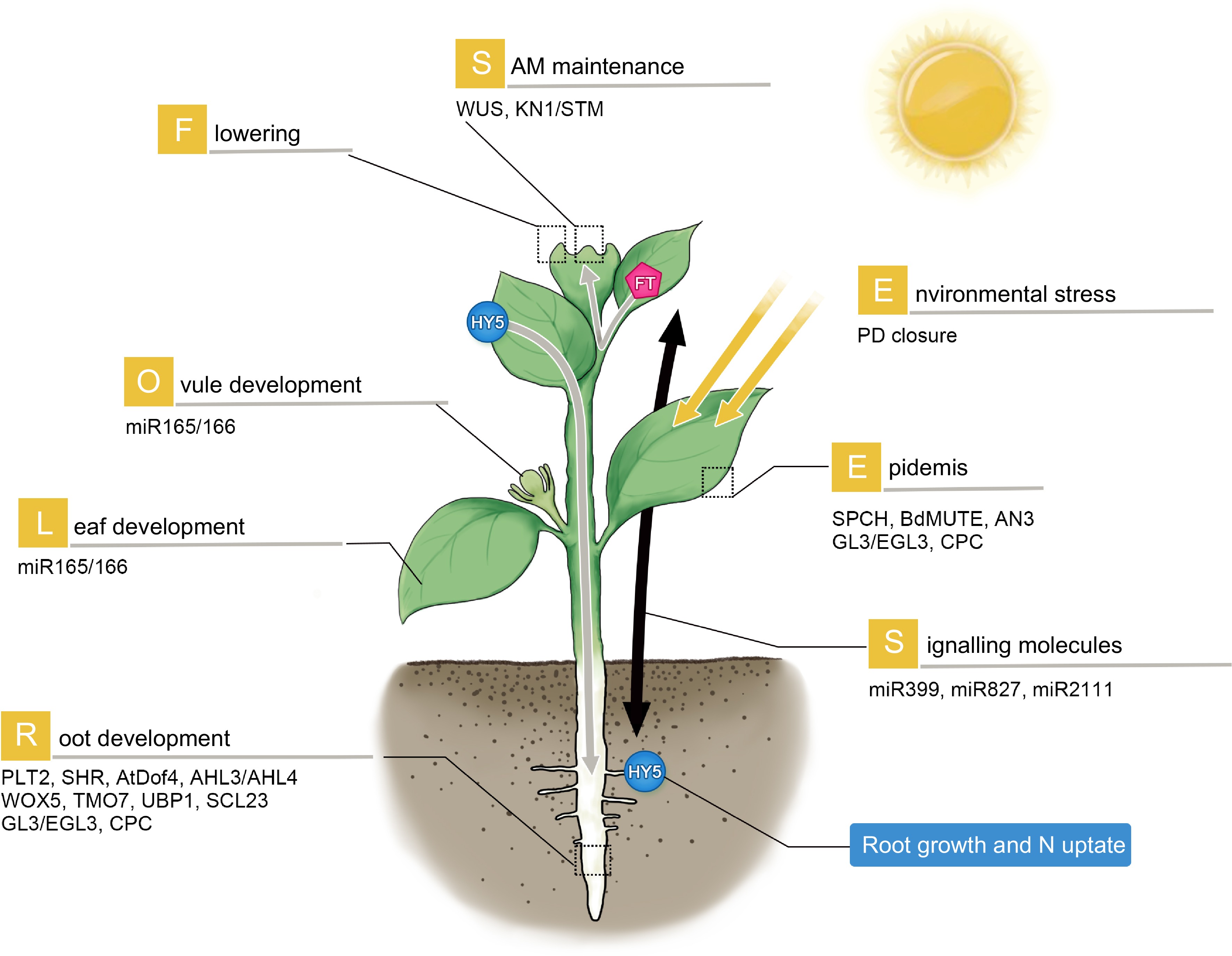
Figure 1.
Mobile proteins and RNAs in plant development and stress response. The mobile regulators participate widely in the development of different organs (as illustrated). They can travel short-range to regulate local tissue patterning or long-distance to transduce systemic signaling. Gray arrow: phloem-based long-distance movement. WUS and STM regulate SAM maintenance; SPCH, BdMUTE, AN3, TTG1, GL3 and CPC are involved in epidermal patterning. In roots, PLT2, SHR, AtDof4.1, AHL3/AHL4, WOX5, TMO7, UBP1 and SCL23 govern a variety of processes including cell division, radial patterning, stem cell maintenance and developmental transition. Long-distance signaling regulators such as FT and HY5 can traffic from leaves to SAM to promote flowering, and from shoot to root to regulate root growth and nitrate uptake respectively. Environmental stresses can induce PD closure. Small RNAs including miR399d, 827 and 2111 move from aerial parts to roots in response to phosphate starvation.
-
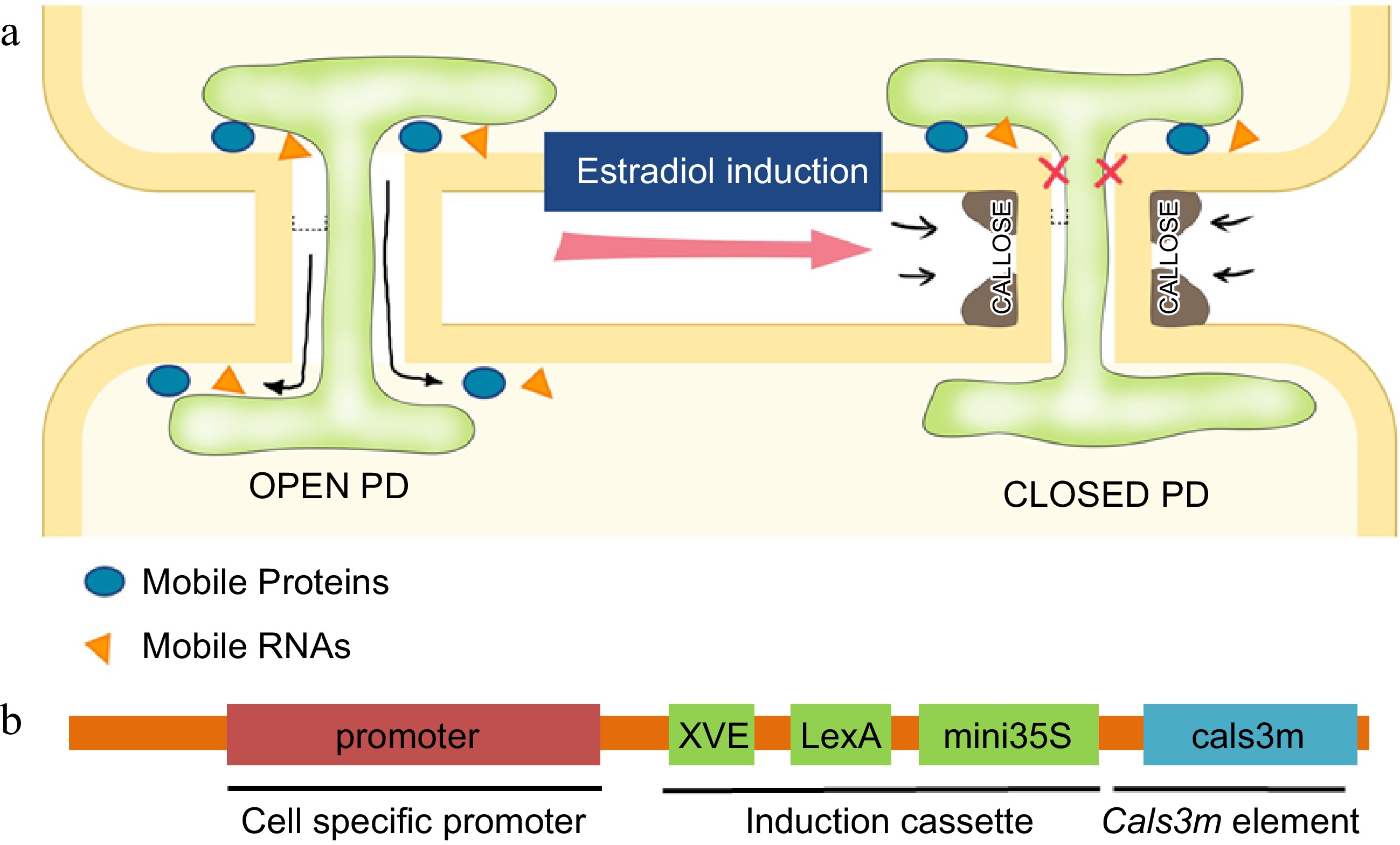
Figure 2.
Regulation of PD permeability by callose. (a) Schematic illustration of regulation of the PD aperture by callose deposition in flanking regions of PD. Induced callose accumulation closes PD permeability and blocks the intercellular movement of transcription factors and small RNAs. (b) The design of ‘icals3m’ system that can inducibly (via estradiol induction cassette) promote callose deposition in specific cell types (via cell-type specific promoters)[128],[138].
-
Mobile TFs Function Moves from:to Reference HY5 Root growth and N uptake Shoot-to-root Chen et al. (2016)[41] DWARF14 Regulate the development of AMs Through phloem into axillary meristems (AMs) Kameoka et al. (2016)[139] BdMUTE BdMUTE is required for subsidiary cell formation GMCs to neighboring cell files Raissig et al. (2017)[97] SPCH Stomatal cell fate Cell-to-cell diffusion in the leaf epidermis of chorus Guseman et al. (2010)[96] AN3 Leaf development From the mesophyll to the epidermis in leaves Kawade et al. (2013)[140] WUS Meristem maintenance From the organizing centre to L1, L2 layers Yadav et al. (2011)[28] KN1/STM Meristem maintenance Broadly in the SAM Kim et al. (2003)[31], 2005[32] PLT2 Longitudinal root zonation Longitudinally from the root meristem forming a gradient Mahonen et al. (2014)[141]; Galinha et al. (2007)[142] SHR Root radial patterning and RAM maintenance Within Stele; Stele into endodermis, QC, CEI and CED Koizumi et al. (2011)[44], Nakajima et al. (2001)[78] AHL3/AHL4 Xylem specification From procambium cells to the xylem Zhou et al. (2013)[37] WOX5 Stem cell maintenance QC to CSC Pi et al. (2015)[30] TMO7 Recruitment of the hypophysis Embryo into the upper cell of suspensor Schlereth (2010)[34]; Lu et al. (2018)[35] Cyp1 Root growth From leaves to root in tomato Spiegelman et al. (2015)[143] UBP1 Transition from cell division to elongation Stele and LRC to cells into transition/elongation zone Tsukagoshi et al. (2010)[144] SCL23 Endodermal cell fate Bidirectional radial spread and movement into meristem Long et al. (2015)[38] TTG1 Trichome patterning Atrichoblasts into trichome initials CPC Trichome patterning, root hair initiation Trichome initials into Atrichoblasts; non-root hair cell into root hair cell Wester et al. (2009)[90] GL3/EGL3 Root hair initiation Root hair cell into non-root hair cell Kang et al. (2013)[91] Table 1.
Summary of the mobile transcription factors identified in plants.
-
Mobile factor Function Moves from: to Reference mRNA KN1 SAM maintenance injected cell to neighbouring cells Lucas et al. (1995)[26] SUC1 Sucrose transport companion cells to sieve elements Kuhn et al. (1999)[145] FT1 Flowering induction Leaf to SAM Lu et al. (2012)[60] Aux/IAA18 Root development Leaf to root Notaguchi et al. (2012)[61] PP16 RNA transport Phloem to shoot apex Xoconostle-Cazares et al. (1999)[62] NACP Meristem maintenance Phloem to shoot apex Ruiz-Medrano et al. (1999)[146] StBEL5 Tuber formation Leaf to root Banerjee et al. (2009)[147] POTH1 Leaf development Leaf to root Mahajan et al. (2012)[148] SLR/IAA14 Lateral root formation Shoot to root Kanehira et al. (2010)[64] PFP-T6 Leaf development Leaf to leaf primordia Kim et al. (2001)[65] PS Pathogen resistance Shoot to root and vice versa Zhang et al. (2018)[149] GAI Leaf development host to parasite Roney et al. (2007) [150]; David-Schwartz et al. (2008)[151] ATC Floral initiation Leaf to flower apices Huang et al. (2012)[152] FVE floral regulators Root to SAM Yang and Yu (2010)[153] AGL24 floral regulators Root to SAM Yang and Yu (2010)[153] siRNA ta-siRNA Establishment of leaf polarity the adaxial to the abaxial side of the leaf Chitwood et al. (2009)[154] hc-siRNA DNA methylation Shoot to root Baldrich et al. (2016)[155] miRNA miR165/166 Xylem specification endodermis into the stele Carlsbecker et al. (2010)[58] miR390 Leaf polarity vasculature and pith region below the SAM to SAM Chitwood et al. (2009)[154] miR394 Meristem maintenance L1 to inner layers in the shoot meristem Knauer et al. (2013)[59] miR395 Sulfate homeostasis graft unions Buhtz et al. (2010)[54] miR399d Phosphate homeostasis shoot to root and vice versa Pant et al. (2008)[156]; Lin et al. (2008)[51] miR172 regulate tuber formation Leaf to root Martin et al. (2009)[55] miR2111 Phosphate homeostasis;
Rhizobial infection;shoot to root and vice versa Huen et al. (2017)[52];
Tsikou et al. (2018)[53]miR827 Phosphate homeostasis shoot to root and vice versa Huen et al. (2017)[52] Table 2.
List of mobile RNAs with functions in organ development.
Figures
(2)
Tables
(2)