-
First visualized by Robert Hooke in 1665, cells had long been regarded as individual units of a whole organism. Whether the cell represents an autonomous entity was a question that had been a subject of debate in 19th Century. The observation of intercellular bridges and plasmodesmata supports the idea that the cellular structure forms the protoplasmic continuity, highlighting the importance of reciprocal interaction of cells within a multicellular organism. As a pioneering cell biologist, Wilson wrote in 1923, "it is the 'organism as a whole' and a 'property of the system as such' "[1], almost all plant cells are connected by the intercellular channel called plasmodesmata (PD)[2].
Primary PD is a straight channel-like structure, as small as 30-50 nm in diameter, connecting two neighboring plant cells[3,4]. A major component of this channel is an endoplasmic reticulum (ER) derived central membranous strands called desmotubles, which form presumably through trapping ER strands in the cell plate during cytokinesis[5,6]. In between the desmotubule and flanking plasma membrane is the cytosolic space called cytoplasmic sleeve[7,8]. Components including cytoskeletons, a GPI-anchor protein and PD localizing proteins (PDLP) have been suggested to participate in the organization and function of plasmodesmata[9, 10].
More recently, sphingolipids were found to affect the pore size of plasmodesmata[11]. Interestingly, analysis of Physcomitrium patens plasmodesmata proteome suggested the enrichment of cell-wall located proteins including EXORDIUM-family members and xyloglucan transglycosylases in plasmodesmata[12]. In particular, this study identified callose-degrading glycolyl hydrolase family 17 (GHL17) proteins as an abundant PD protein family[12], suggesting the potentially conserved plasmodesmata regulation by callose (will be further discussed later in this review) over the evolution.
Smaller molecules, ions and metabolic substance can all pass through PD by diffusion. Other micro-molecules including proteins and RNAs are thought to transverse PD via active transport[11−15]. Mobile molecules can move across PD via either the cytoplasmic sleeve, or through the desmotubule (in lumen or lateral diffusion in the desmotubule membrane), or via diffusion in the flanking plasma membrane[16,17]. In support of these hypotheses, it was found that the interference of the membrane structure affected PD permeability[17]. In old tissues, plant cells further produce secondary PD that is normally branched and complex in shape. Localized cell wall modification could be involved in secondary PD formation, and the complexity of this type of PD is correlated with reduced PD permeability[18,19]. Nevertheless,the detailed mechanism and the exact roles of secondary PD during development are still far from clear. Interestingly, multiple types of PD were found at grafted wounds, suggesting that different PD types could have distinct functions[20]. In this review, we focus on our current understanding of cell-to-cell signaling across plasmodesmata.
-
The observation of cell-to-cell movement of large molecules initially arose from the micro-injection of fluorescent dye in plant tissues[21−24]. The first endogenous protein exhibiting the intercellular mobility is KNOTTED1 (KN1), a homeodomain protein essential for maintenance of the shoot apical meristem (SAM) in maize[25,26]. Recently, the ribosomal RNA-processing protein 44A (AtRRP44A) was shown to mediate the cell-to-cell trafficking of KN1[27]. Since then, a large number of transcription factors were identified in plants that can move between tissues and cells to provide positional instruction during plant development[21]. These mobile regulators can traffic across just a few cell layers to function locally or over a long distance to affect global developmental change.
One of the central questions in organogenesis is how to spatiotemporally maintain stem cells and specify cell fates. In SAM, WUSCHEL (WUS) is expressed in the organizing center of shoot apical meristem, but the protein moves to the layer1 and 2 (L1 & 2) of shoot apical meristem where WUS triggers CLAVATA 3 (CLV3) expression, which in turn inhibits WUS transcription in L1 and L2 layer[28,29]. With this WUS-CLV3 feedback loop, plants can maintain the stem cell population in proper size in SAM. With the similar strategy, plants maintain the root stem cell niche via WOX5-CLE40 loop, in which WOX5 traffics from quiescent center (QC) to columella stem cell (CSC) to repress the cell differentiation[30]. In Arabidopsis, SHOOT MERISTEMLESS (STM) and ARABIDOPSIS KNOTTED-LIKE (KNAT1)/BREVIPEDICELLUS (BP) are two homologs of the KN1 gene, previously described to be mobile in maize SAM. When driven by an L1 specific promoter, STM and KNAT1 were observed to move from the L1 layer into the inner cell layers of the SAM[31,32]. In addition, KNAT1 was able to pass the interface between cortex and epidermis in Arabidopsis when mis-expressed by a mesophyll specific promoter[33].
In embryogenesis, TARGET OF MONOPTEROS 7 (TMO7), encoding a bHLH transcription factor, is essential for hypophysis, the founder cell for forming root apex during post-embryonic growth. TMO7 is transcribed in embryonic cells while the TMO7-GFP fusion can be detected in the neighboring hypophysis, indicating a non-cell-autonomy of this regulator[34,35]. In post-embryonic growth, intercellular movement of transcriptional factors regulates a variety of developmental aspects ranging from root radial patterning to root hair and trichome initiation. These mobile regulators including SHORT-ROOT (SHR), CAPRICE (CPC), TRANSPARENT TESTA GLABRA 1 (TTG1), GLABRA 3 (GL3), ENHANCER OF TRY AND CPC 3 (ETC3)/ TRIPTYCHON (TRY), UBIQUITIN-SPECIFIC PROTEASE (UBP1) have been well reviewed previously[15,21]. A previous screen estimated that around 15% of transcriptional factors in roots can move between cells[36]. In contrast, we only have limited understanding of the functionality of these mobile proteins.
Recently, more mobile transcriptional factors have been identified (summarized in Table 1). Two closely related AT-hook family members, AT-HOOK MOTIF NUCLEAR LOCALIZED PROTEIN 3 (AHL3) and AHL4, were shown to interact in vivo and regulate the boundaries between the procambium and xylem[37]. Interestingly, their interaction seemed to be required for their intercellular trafficking. A SHR target, SCL23 displays a bidirectional radial spread and long-range movement into meristem in Arabidopsis roots. Through direct interaction, SCL23 controls movement of SHR and participate in endodermal specification in the root meristem[38].
Table 1. Summary of the mobile transcription factors identified in plants.
Mobile TFs Function Moves from:to Reference HY5 Root growth and N uptake Shoot-to-root Chen et al. (2016)[41] DWARF14 Regulate the development of AMs Through phloem into axillary meristems (AMs) Kameoka et al. (2016)[139] BdMUTE BdMUTE is required for subsidiary cell formation GMCs to neighboring cell files Raissig et al. (2017)[97] SPCH Stomatal cell fate Cell-to-cell diffusion in the leaf epidermis of chorus Guseman et al. (2010)[96] AN3 Leaf development From the mesophyll to the epidermis in leaves Kawade et al. (2013)[140] WUS Meristem maintenance From the organizing centre to L1, L2 layers Yadav et al. (2011)[28] KN1/STM Meristem maintenance Broadly in the SAM Kim et al. (2003)[31], 2005[32] PLT2 Longitudinal root zonation Longitudinally from the root meristem forming a gradient Mahonen et al. (2014)[141]; Galinha et al. (2007)[142] SHR Root radial patterning and RAM maintenance Within Stele; Stele into endodermis, QC, CEI and CED Koizumi et al. (2011)[44], Nakajima et al. (2001)[78] AHL3/AHL4 Xylem specification From procambium cells to the xylem Zhou et al. (2013)[37] WOX5 Stem cell maintenance QC to CSC Pi et al. (2015)[30] TMO7 Recruitment of the hypophysis Embryo into the upper cell of suspensor Schlereth (2010)[34]; Lu et al. (2018)[35] Cyp1 Root growth From leaves to root in tomato Spiegelman et al. (2015)[143] UBP1 Transition from cell division to elongation Stele and LRC to cells into transition/elongation zone Tsukagoshi et al. (2010)[144] SCL23 Endodermal cell fate Bidirectional radial spread and movement into meristem Long et al. (2015)[38] TTG1 Trichome patterning Atrichoblasts into trichome initials CPC Trichome patterning, root hair initiation Trichome initials into Atrichoblasts; non-root hair cell into root hair cell Wester et al. (2009)[90] GL3/EGL3 Root hair initiation Root hair cell into non-root hair cell Kang et al. (2013)[91] Besides the local regulation, transcriptional factors were also found to traffic long-distance between organs to direct global developmental transition in plants in Fig 1. An early example is the detection of FLOWERING LOCUS T (FT) trafficking from leaves where it is synthesized in response to day length, to the
SAM to trigger flowering[39,40]. Recently, a light-activated transcriptional factor, ELONGATED HYPOCOTYL 5 (HY5) was shown to move via phloem from shoot-to-root. This translocation of HY5 was proposed to mediate light-activated root growth and N uptake from the soil to balance photosynthetic carbon fixation in the leaf[41]. 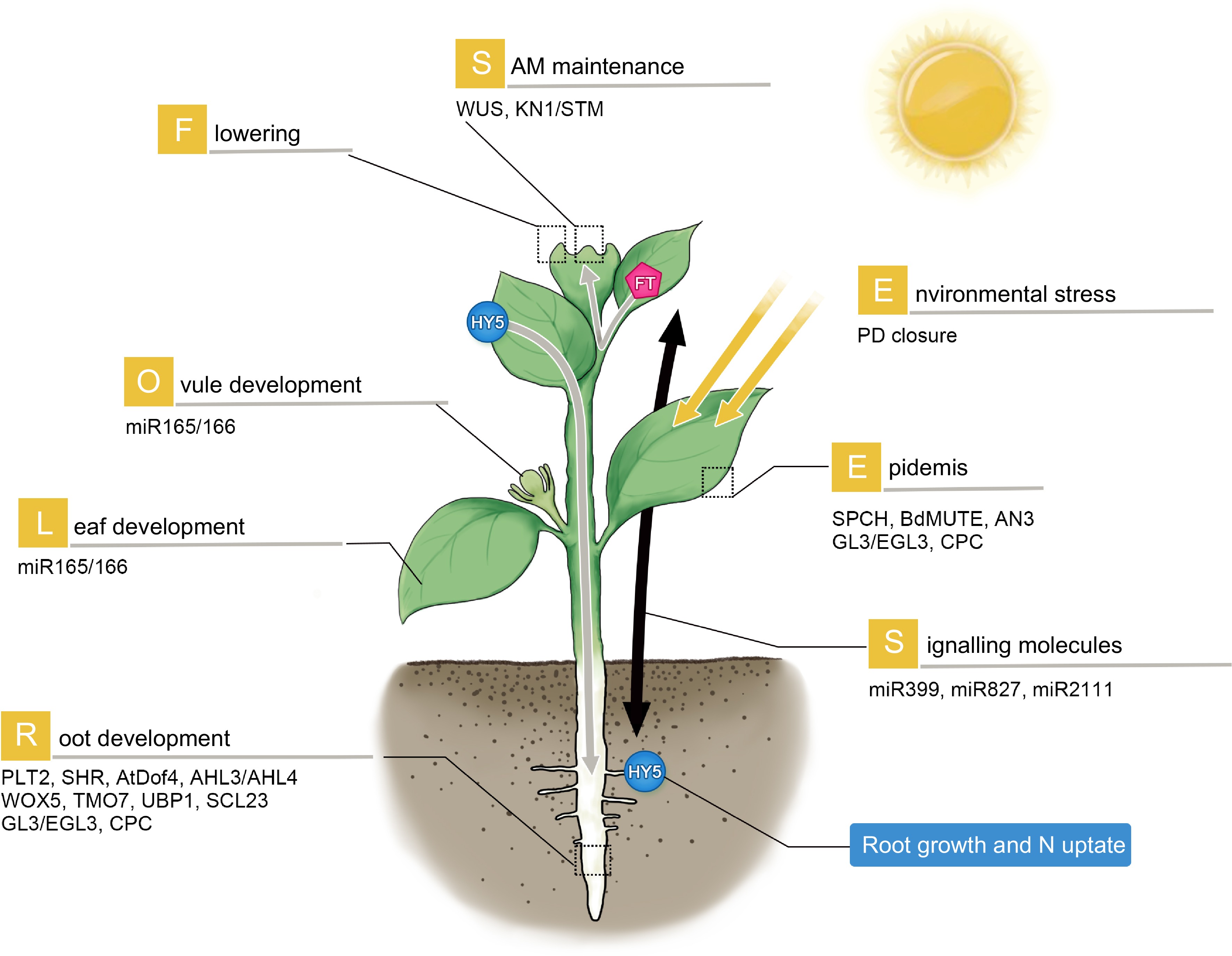
Figure 1.
Mobile proteins and RNAs in plant development and stress response. The mobile regulators participate widely in the development of different organs (as illustrated). They can travel short-range to regulate local tissue patterning or long-distance to transduce systemic signaling. Gray arrow: phloem-based long-distance movement. WUS and STM regulate SAM maintenance; SPCH, BdMUTE, AN3, TTG1, GL3 and CPC are involved in epidermal patterning. In roots, PLT2, SHR, AtDof4.1, AHL3/AHL4, WOX5, TMO7, UBP1 and SCL23 govern a variety of processes including cell division, radial patterning, stem cell maintenance and developmental transition. Long-distance signaling regulators such as FT and HY5 can traffic from leaves to SAM to promote flowering, and from shoot to root to regulate root growth and nitrate uptake respectively. Environmental stresses can induce PD closure. Small RNAs including miR399d, 827 and 2111 move from aerial parts to roots in response to phosphate starvation.
Considering the size of transcriptional factors, PD seems to be the most possible way for the intercellular translocation. With an iclas3m system (described in detail in a later part of this review) that blocked the PD between stele and endodermis, SHR intercellular transport was terminated[3]. Another piece of evidence supporting PD transport of transcriptional factors is the blocked movement of TMO7 from meristematic cells into the root cap in the cals3-2d, a mutant in which PD is restricted by over-accumulated callose[35]. To get access to PD, transcriptional factors could exploit intracellular apparatus including microtubules and endomembrane delivery system[42,43]. Besides, an unknown function protein named SHR INTERACTING EMBRYONIC LETHAL (SIEL) was shown to interact with a number of mobile transcriptional factors and the mutation of this gene seemed to reduce SHR intercellular movement[44]. As SIEL partially localized to endosomes, it was proposed that this protein could function as a 'shuttle' to facilitate delivery of mobile transcriptional factors. In addition, some facilitating proteins have also been identified. After passing through PD, a few mobile proteins including APS KINASE 1 (KN1), SHOOT MERISTEMLESS (STM) and TRANSPARENT TESTA GLABRA 1 (TTG1) were discovered to associate with a group of type II chaperonin complexes consisting of CHAPERONIN CONTAINING T-COMPLEX POLYPEPTIDE-1 SUBUNIT 7 and 8 (CCT7 & CCT8), which facilitate the movement possibly by promoting the protein refolding after the PD cross-over[27].
Although no specific domain has been identified that accounts for intercellular mobility, the cell-to-cell transport of transcriptional factors seemed to be protein sequence-dependent. Homeodomain (HD) and the helical domains have been shown to be necessary and sufficient for PD-mediated transport of KN1. Unlike this, three conserved domains (HD, WUS-box, and EAR-like domain) in WUS
are not required for its movement. Instead, WUS mobility seems to be controlled by a non-conserved sequence between the HD domain and WUS-box[29]. Despite triple GFP Tag impaired TMO7 movement, protein size did not seem to be the primary determinant of intercellular transport. Instead, TMO7 was found to move in a sequence-dependent manner, and both nuclear residence and protein modification are important for TMO7 mobility[35]. In two other mobile transcriptional factors, CPC and SHR, the mobility relied on multiple regions within the proteins. In addition, the mobility of these two proteins seemed to be associated with the subcellular distribution in both the cytoplasm and the nucleus. In addition to transcriptional factors, small RNAs also participate in transcriptional regulation of diverse developmental and physiological events in plants. Small RNAs are 21−24 nt long and can be generally divided into siRNAs and miRNAs[45]. Small RNAs function either through degrading target genes by near-perfect complementarity, or via transcriptional silencing by histone modification and DNA methylation[46−50]. Small RNAs were often regarded as the long-distance signals as the initial efforts dissecting their mobility exploited the grafting system in which mutants defective in small RNAs biogenesis were included. Facilitated by high-throughput sequencing techniques, researchers identified a large number of mobile siRNAs that can traffic from shoot to root presumably via phloem. Besides siRNA, a large number of miRNAs were discovered to traffic in phloem exudates over long distance. Low-phosphate induced miR399s exhibited a shoot-to-root movement to repress downstream targets including PHO2 in the root[51]. Similarly,miR399d, miR827 and miR2111 were all found in grafting experiments to relocate from aerial parts to roots in response to phosphate starvation[52]. During rhizobial infection, miR2111 functioned as long-distance signals to post-transcriptionally regulate symbiosis suppressor TOO MUCH LOVE in roots[53]. miR395 can also translocate from wild-type scions to rootstocks of the miRNA processing mutant hen1-1 to target the APS gene[54]. In addition, both miR156 and miR172 have been confirmed as potentially phloem-mobile miRNAs that regulate tuber formation[55−57].
In grafting system, only small RNAs transporting from shoot-to-root via phloem could be analyzed. Other approaches that allow for the comparison between the expression areas and in situ RNA distribution patterns may help the identification of small RNAs acting locally as non-cell autonomous signals. To establish adaxial–abaxial leaf polarity, a member of Trans-acting small interfering RNA (ta-siRNA) family forms a gradient across the leaves by intercellular diffusion. This diffusion-driven pattern of ta-siRNA shapes the expression pattern of AUXIN RESPONSE FACTOR3 (ARF3), an abaxial determinant gene. Another small RNA, miR390 was proved to regulate the leaf polarity by the cell-to-cell movement from vasculature and pith region below the shoot apical meristem to the vegetative apex[54]. In addition, miRNA165/166 were discovered to move from the endodermis into the stele to regulate the xylem cell fate[58]. Moreover, miR394 was shown to regulate stem cell maintenance in SAM by the PD-mediated movement from L1 to inner cell layers to repress LEAF CURLING RESPONSIVENESS (LCR) expression[59].
In addition to siRNA and miRNA, mRNAs have also been found to travel beyond the cells in which they are expressed in Fig 1. In addition to the early example of mobile mRNAs of KN1, potato sucrose transporter SUC1 mRNA was also confirmed to be mobile. In grafting experiments, a number of mRNAs were found to travel, such as FT, FVE and AGL24 in Arabidopsis[60], Aux/IAA in melon and Arabidopsis[61], PP16 and NACP in pumpkin[62,63], BEL5 and POTH1 in potato, SLR/IAA14 in apple[64], PFP-T6 and PS in tomato[65] (summarized in Table 2). Recently, Luo et al. developed a fluorescence-based mRNA labeling system to identify mobile mRNAs targeted to PD[66]. Their analyses revealed that only mobile rather than not non-mobile mRNAs were selectively targeted to PD, providing further evidence for PD mediated transport of mRNAs. Interestingly, using a Nicotiana benthamiana/tomato heterograft system, Xia et al. found some mRNAs have bidirectional mobility between shoots and roots. In addition, forced expression of non-mobile mRNAs in the companion cells did not confer the mobility[67−71]. Thus, the movement of mRNA is likely an actively regulated process. Moreover, a large number of graft-transmissible mRNAs have been identified by high throughput sequencing in a variety of species including Arabidopsis, tobacco, grape, cucumber and tomato[67−72].
Table 2. List of mobile RNAs with functions in organ development.
Mobile factor Function Moves from: to Reference mRNA KN1 SAM maintenance injected cell to neighbouring cells Lucas et al. (1995)[26] SUC1 Sucrose transport companion cells to sieve elements Kuhn et al. (1999)[145] FT1 Flowering induction Leaf to SAM Lu et al. (2012)[60] Aux/IAA18 Root development Leaf to root Notaguchi et al. (2012)[61] PP16 RNA transport Phloem to shoot apex Xoconostle-Cazares et al. (1999)[62] NACP Meristem maintenance Phloem to shoot apex Ruiz-Medrano et al. (1999)[146] StBEL5 Tuber formation Leaf to root Banerjee et al. (2009)[147] POTH1 Leaf development Leaf to root Mahajan et al. (2012)[148] SLR/IAA14 Lateral root formation Shoot to root Kanehira et al. (2010)[64] PFP-T6 Leaf development Leaf to leaf primordia Kim et al. (2001)[65] PS Pathogen resistance Shoot to root and vice versa Zhang et al. (2018)[149] GAI Leaf development host to parasite Roney et al. (2007) [150]; David-Schwartz et al. (2008)[151] ATC Floral initiation Leaf to flower apices Huang et al. (2012)[152] FVE floral regulators Root to SAM Yang and Yu (2010)[153] AGL24 floral regulators Root to SAM Yang and Yu (2010)[153] siRNA ta-siRNA Establishment of leaf polarity the adaxial to the abaxial side of the leaf Chitwood et al. (2009)[154] hc-siRNA DNA methylation Shoot to root Baldrich et al. (2016)[155] miRNA miR165/166 Xylem specification endodermis into the stele Carlsbecker et al. (2010)[58] miR390 Leaf polarity vasculature and pith region below the SAM to SAM Chitwood et al. (2009)[154] miR394 Meristem maintenance L1 to inner layers in the shoot meristem Knauer et al. (2013)[59] miR395 Sulfate homeostasis graft unions Buhtz et al. (2010)[54] miR399d Phosphate homeostasis shoot to root and vice versa Pant et al. (2008)[156]; Lin et al. (2008)[51] miR172 regulate tuber formation Leaf to root Martin et al. (2009)[55] miR2111 Phosphate homeostasis;
Rhizobial infection;shoot to root and vice versa Huen et al. (2017)[52];
Tsikou et al. (2018)[53]miR827 Phosphate homeostasis shoot to root and vice versa Huen et al. (2017)[52] -
A plant organ is usually composed of morphologically and functionally different cell types in different positions. Small molecules can move between cells and across plasmodesmata, which mediates crucial intercellular communication for the growth and development of plant tissues and organs. For example, a plant root is composed of concentrically arranged cell layers with epidermis, cortex, endodermis, and stele locating from outside to inside[73]. This anatomic arrangement highlights the regulation of tissue patterning instructed by positional information, often through the exchange of signaling molecules between cells. A number of developmental processes including root radial patterning, root hair initiation and trichome formation, have emerged as the model system for studying tissue patterning in plants.
In root, the formation of the endodermal cell layer starts from the endodermal and cortex initial cells in root stem cell niche, where two transcriptional factors, SHR and SCARECROW (SCR) promote the expression of CYCD6;1 to allow the switch of cell division pattern from anticlinal to periclinal[74−77]. This results in the formation of two distinct layers of cells within the ground tissue, and the role of SHR in specifying the endodermal layer was proposed based on the fact that the endodermal layer was completely absent in shr-2 mutant. Intriguingly, SHR expression is restricted in stele, but the SHR protein is actively transported through PD from stele toward the outside to play non-cell-autonomous roles[78,79]. In the enodermis, SHR directly activates SCR which, in turn, physically binds to SHR to trap this mobile transcription factor in the nucleus of the endodermis, preventing further movement[77]. This mechanism was discovered to be conserved in rice and thus was proposed to be an evolutionarily conserved mechanism defining a single endodermal cell layer in almost all land plants[74]. However, a study on rice SHR homologs suggested that SHR alone is insufficient to determine endodermal cell fate[80]. Consistent with this argument, mis-expression of SHR indicated that SHR ability to confer endodermal identity partially relied on cell lineage and was coordinated by uncharacterized positional information, presumably derived from stele.
Specific expression of marker genes, as often used previously to determine endodermal cell fate, is sometimes misleading. A prominent feature of the endodermis is the formation of the Casparian Strip (CS), an apoplastic barrier between vascular tissues and outer ground tissues[81]. The presence of functional CS is therefore a better trait for precise evaluation of endodermal identity. Two recent studies revealed that SHR does serves as a master regulator activating a hierarchical downstream network for CS formation[82,83]. The combination of SHR mediated cascade and another independent peptide signal derived from stele forms the minimum set of regulators that program endodermal identity, exemplified by the formation of functional CS[83]. Since both SHR and the peptide are specifically expressed in vascular tissues, CS formation represents the elaborate developmental control by stele-to-endodermis movement of mobile regulators. Besides CS, SHR and its downstream target SCR can activate the expression of miRNA165/166 in the endodermis which in turn moves back to vasculature to repress a class III homeodomain-leucine zipper transcription factors for proper xylem formation[58]. Thus the reciprocal communication between ground tissue and vasculature in root spatially defines the radial patterning in root. In Cardamine, a recent study indicated that a differential spatial distribution of miR165/166 is responsible for forming the extra cortex layer[84]. In addition to roots, miR165/166 also function in other organs including leaf primordial and ovule. By restricting PHB expression in incipient inner integument, miR165/166 promotes the correct ovule patterning[85]. Interestingly, a callose synthase mutant in maize, named tie-dyed2 (tdy-2), affects the development of vasculature, suggesting the mechanism of vascular development directed by intercellular communication (possibly via miR165/166) is likely conserved in crops[86,87]. In addition to roots, plasmodesmata also plays a key role in regulating leaf development, particularly the formation of leaf veins[88].
Trichomes and root hairs, originating from the epidermis in leaves and roots respectively play important roles in protecting plants from bio/abiotic stresses, and promoting nutrient absorption[89,90]. In Arabidopsis, the initiation of trichomes and root hairs is precisely patterned in epidermis, indicating an essential role of cell-to-cell communication in these processes.
In trichome initiation, both positive regulator TRANSPARENT TESTA GLABRA (TTG1) and negative regulator ENHANCER OF TRY AND CPC 3 (ETC3) and CAPRICE (CPC) move between cells. In incipient trichome cells, TTG1 protein accumulates through a trapping/depletion mechanism mediated by GLABRA3 (GL3)[91]. On the other hand, the repressor of ETC3 and CPC move into the neighboring non-trichome cells (also regulated by GL3), forming inactivated MYB/bHLH/WD40 to inhibit the development towards trichomes[92]. Recently, PdBG4 has been implicated in regulating PD permeability in Arabidopsis trichome development[93]. In root hairs, CPC serves as a positive regulator and it is trapped in the hair-position root epidermis by interacting with EGL3 and GL3 after the movement[94]. The trn1 mutant is defective in the position-dependent pattern of root hairs and cause the ectopic expression of WER, GL2 and EGL3, suggesting that TRN1 also participates in the position-dependent cell fate determination[95,96].
Stomata on epidermis are responsible for water and gas exchange between the plants and the environments. The mature stomata structure is produced through successive cell division and differentiation process, with both processes subject to highly spatiotemporal regulation[97]. In a GLUCAN SYNTHASE-LIKE 8 (GSL8) mutant in which normal callose deposition is disrupted, SPCH-GFP diffused to neighboring cells from meristemoids, resulting in excessive proliferation of stomatal-lineage cells. This observation suggests that proper gating of critical regulators, likely through callose regulation, regulates the correct patterning of stomata complex[98]. MUTE, another key transcriptional factor required to terminate asymmetric division and promote the transition of meristemoids to GMCs, was shown in Brachypodium to move from GMCs to neighboring cells to induce the subsidiary cells (SCs) formation[99].
-
Plants respond to stresses often by accumulation of callose, which is negatively correlated with PD permeability in Fig 2. A variety of abiotic stresses have been associated to callose induction, such as cold stress[100,101], wounding[102,103], heat stress[104,105], and heavy metals[106−109]. Although detailed mechanism is not entirely clear, callose synthases were found to participate in the callose regulation. In Arabidopsis, there are 12 callose synthase (CalS) family members. When exposed to excess iron, the cals5 and cals12 mutants showed an attenuated callose deposition in phloem, compared to wild type and other cals mutants. This result suggests that cals5 and cals12 may play specific roles in iron stress response in Arabidopsis[110]. In tomato, cold stress has long been known to cause catfacing fruits or malformed fruits by breeders and gardeners. A recent study proved this phenomenon was caused by the restriction of SlWUS intercellular movement via plasmodesmata in floral meristem[101]. The cold induced callose accumulation blocked the plasmodesmata, resulting in the excessive activation of CLV3 and TAG1, and disrupted WUS-CLV3/WUS-TAG1 negative feedback loops[101].
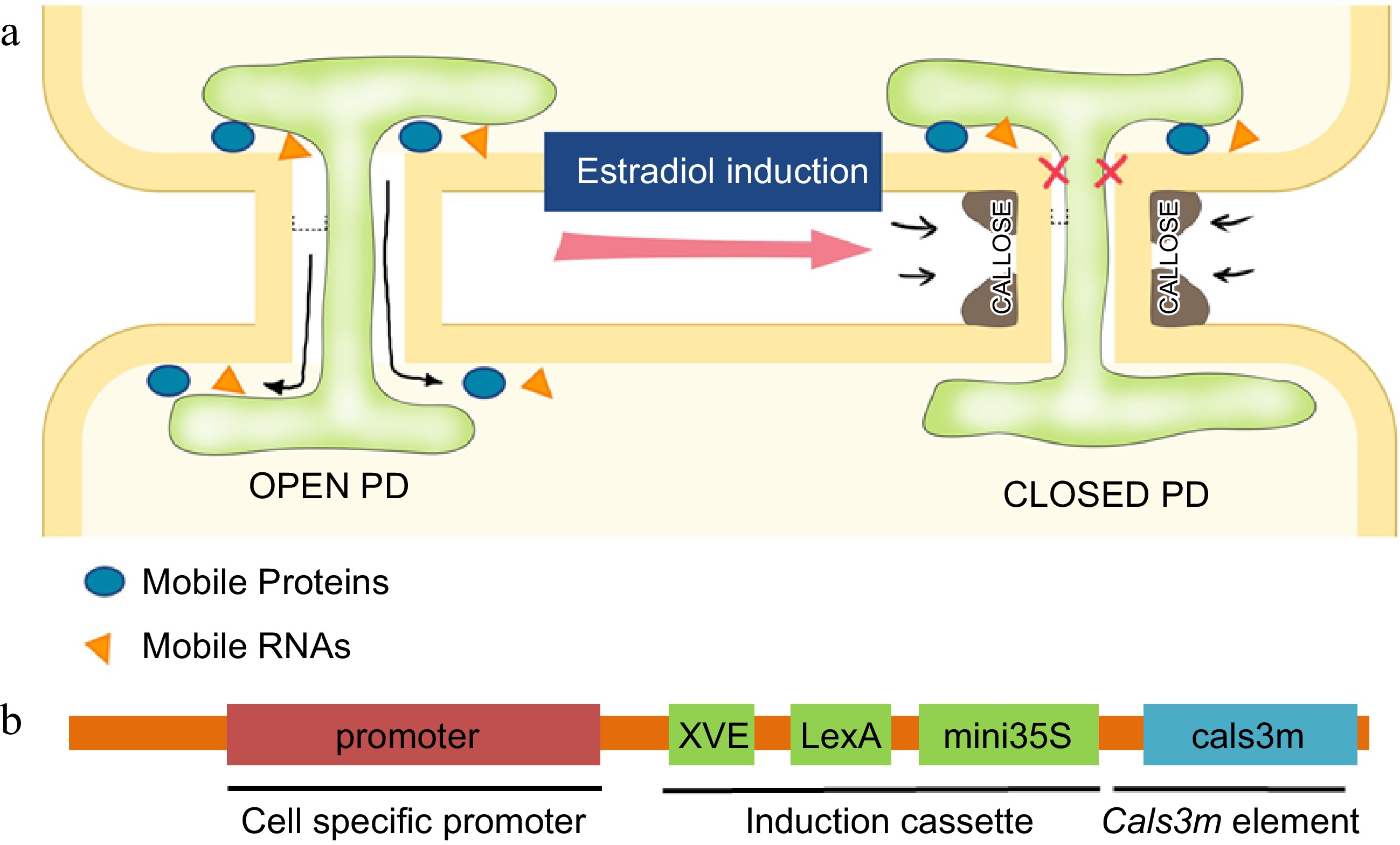
Figure 2.
Regulation of PD permeability by callose. (a) Schematic illustration of regulation of the PD aperture by callose deposition in flanking regions of PD. Induced callose accumulation closes PD permeability and blocks the intercellular movement of transcription factors and small RNAs. (b) The design of ‘icals3m’ system that can inducibly (via estradiol induction cassette) promote callose deposition in specific cell types (via cell-type specific promoters)[128],[138].
It has been reported that PD regulation serves as an innate defense strategy[111]. Pathogens trigger both pathogen-associated molecular pattern (PAMP) and PAMP-triggered immunity (PTI) systems, which have been reported to induce callose deposition[112]. Upon SMV virus invasion, callose was accumulated in soybean phloem which prevents the virus from traveling long distances[113]. Salicylic acid (SA) is a plant immune signal produced upon pathogen infection, which has also been shown to trigger PD closure and affect symplastic communication. Elevation of SA level seemed to be necessary for the PD response during bacterial infection, and the expression of bacterial derived salicylate hydroxylase (NahG) gene in plants resulted in higher susceptivity to bacteria[113]. Biotic stresses including pathogen infection are known to modulate ROS level and callose abundance in infected regions, which is presumably responsible for the altered PD permeability[114,115].
Virus can also regulate the mesenchymal plasmodesmata in tobacco[109] and it was recently reported that ROS-mediated PD closure is controlled by multiple pathways, either in SA- or PDLP5-dependent manners. Change of callose level in biotic stresses is also modulated by callose synthase members[112,113]. SA-dependent PD regulation requires the function of callose synthase1 (CalS1). However, the CalS8 seemed to be more involved in basal and ROS-dependent PD regulation[103]. Callose synthase members have also been widely reported in recent years. CsCalS4 function was identified in pollen development in cucumber, and CsCalS1/8 homologous genes were induced by cucumber fungus and functioned as the key factors in response to biological stress[114]. GhCalS5 and ZmCals were found to promote callose synthesis in cotton and maize in responsive to stresses[116,117].
In addition, PD-localized proteins also emerged as the regulator of PD aperture during biotic stresses. It was shown that the PD closure triggered by chitin was dependent on the activity of PD-localized receptor-like protein LYM2[111]. Besides, bacterial flagellin could rapidly activate the expression of CML41, a PD-localized Ca2+-binding protein, which is necessary for the induction of callose at PD.
Callose is the linear polysaccharide that is composed of β-1,3-glucan. It is a component of cell wall and is frequently found to deposit at PD, where it is believed to control the PD permeability during plant development and stress response. It was found the precise developmental transition often relies on the regulation of symplastic continuity. In birch, bud dormancy entry and release are associated with the shift between callose production and turnover. Callose accumulation at PD in the shoot apical and rib meristems can seal off the symplastic communication and promote the bud dormancy[116−121]. A period of chilling, however, triggers gibberellin biosynthesis, resulting in increased expression of 1,3-β-glucanases and degradation of callose. Accumulating evidence suggests that callose regulation is actually implicated in a wide range of developmental processes, including seed germination, embryogenesis, cell division, flowering and reproduction[122−124]. In tomato, a short period of cold stress is sufficient to induce callose accumulation in floral meristem and blocked intercellular movement of SlWUS, resulting in malformed fruits[101]. In olives, callose deposition, as part of cell wall modification, regulates fruit abscission[114].
Through a genetic screen for defective vascular development, Vaten et al. (Helariutta group) identified three semi-dominant alleles of CALLOSE SYNTHASE 3 (cals3d) that caused an increase in callose deposition at PD and abnormal plant growth[3,19]. In the root, cals3d mutants all showed aberrant radial patterning and misspecification of the phloem and the xylem. Consistent with these phenotypes, cals3d roots exhibited decreased PD-mediated symplastic movement of free GFP, SHR and miRNA165/66[3,125]. It thus seemed that the identified dominant mutations can substantially enhance the ability of CALS3 to promote callose deposition at PD. By combining these mutations in a vector containing LexA-VP16-ER (XVE)-based estradiol inducible cassette, the Helariutta group designed an elegant tool named as the 'icals3m system'. Driven by specific promoters, this system can potentially be used to temporally manipulate callose at PD and symplastic communication in particular cell types[3,126].
The initial attempts using this system in vascular tissues and lateral root development proved to be successful[3,125,127]. With specific induction of icals3m system in xylem pole pericycle, Benitez-Alfonso et al. detected a significantly increased number of initiated primordial [126]. Together with the observation of a transient symplastic isolation of the primordium prior to emergence, they confirmed the essential role of callose based symplastic connectivity between pericycle cells, founder cells, and the neighboring tissue during lateral root patterning[122]. More recently, icals3m system was used to dissect the roles of symplastic communication in root apical stem[122]. Driven by an endodermis-specific EN7 promoter, icals3m induced symplastic blockage led to severe root patterning defects, shown by disrupted cell division direction, misspecification of cell fate as well as impaired cell polarity. In root tip, different cell types including endodermis all derived from the root stem cell niche, where QC was believed to repress the differentiation of surrounding stem cells based on an early classic laser ablation experiment carried out in the 1990's[127]. However, icals3m system provides an alternative non-invasive approach to examine the role of QC. With the expression under WOX5 promoter, icals3m system was clearly shown to induce callose specifically in QC[128]. The visible callose signal based on aniline blue staining was detected as quickly as 6 h after the estradiol induction[129]. This icals3m system was further used to study the interaction between root cap and the root meristem[124,128−130]. When the symplastic communication between root cap and root meristem was disrupted, developmental defects were observed in both parts: In meristem, stem cell maintenance was affected while in root cap the starch granules, the marker commonly used as an indicator of columella differentiation, disppeared[125]. An earlier study showed that starch granules in columella cells relied on auxin concentration[131]. In this study, short-term disruption of symplastic communication was sufficient to cause defects in stem cells, while it took longer for auxin distribution in root meristem to occur[125]. In fact, plasmodesmata itself can act as the channel for auxin flow[131,132]. Furthermore, icals3m system also was employed in the study of phloem unloading[132]. A phloem pole pericycle specific promoter CalS8 and a companion cell and metaphloem sieve element specific promoter psAPL were both used to drive icals3m to block the connection between different phloem cell types[133]. A direct developmental defects arose from the blocked plasmodesmata in phloem was the reduced growth of axillary buds[50].
To summarize, callose regulation is a central mechanism to control symlastic communication during plant development. Spatiotemporal expression of icals3m system can be an effective tool to deepen our understanding of the developmental regulation mediated by symplastic signals. The power of this system can be even higher with the combination with other techniques including cell type specific OMICs. The application of this system in vegetable studies would greatly enhance our ability to dissect various aspects of development and physiology in vegetable species ranging from fruit development to stress resistance.
-
Intercellular signaling across plasmodesmata plays crucial roles in a wide range of processes in plants. The currently identified signaling molecules across plasmodesmata are mainly transcription factors and RNAs. However, accumulating evidence suggests that many other signaling pathways including calcium signaling, redox signaling, phosphorylation signaling, and hormone signaling can also function in non-cell-autonamous manner[134−136]. As these pathways are often complex and interplay with each other, it is still difficult to unravel such non-cell-autonamous functions. With the advance in high-resolution imaging techniques, such as super-resolution microscopy, researchers will be able to visualize in vivo the action and mobility of the molecular players involved in intercellular signaling[137].
In addition to visualizing the intercellular mobility of molecules, it is crucial to precisely evaluate the phenotype with a specific intercellular signaling disrupted. Developing cell type specific approaches is the key step and thus identification of promoters with restricted expression in certain cell types is important. Furthermore, abolishing gene function in a specific cell type is a valuable tool for studying intercellular signaling. Previously, cell-specific RNAi was employed but the intercellular mobility of small RNAs prevents the precise evaluation of gene function. Recent rapid development of CRISPR-Cas9 technique has emerged as a powerful tool for this purpose. The combination of cell-specific expression of Cas9 with reporters that allows for visualizing the gene editing in different cells could greatly enhance our ability to precisely evaluate the function of mobile regulators.
Lastly, to gain a more comprehensive understanding of the plasmodesmata mediated intercellular signaling, it is important to integrate multiple approaches, such as high-resolution imaging, single-cell technique, multi-omics, and computational modeling. Although the cell-to-cell signaling often occurs locally, the impact could be systemic in plants. The complete assessment of plasmodesmata-mediated intercellular signalling, as well as derived tissue- or cell-type-specific techniques, will not only benefit the study of plant development, but also provide the opportunity for future biotechnological renovation of plants.
This work was supported by carbon-nitrogen high efficiency grants from Fujian Agriculture and Forestry University (118992201A).
-
The authors declare that they have no conflict of interest.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Li M, Niu X, Li S, Li Q, Fu S, et al. 2023. Intercellular signaling across plasmodesmata in vegetable species. Vegetable Research 3:22 doi: 10.48130/VR-2023-0022
Intercellular signaling across plasmodesmata in vegetable species
- Received: 20 December 2022
- Accepted: 10 May 2023
- Published online: 02 August 2023
Abstract: The formation of edible organs and stress adaption are two major focuses of the studies on vegetable species. The regulation of these two processes often involves cell-to-cell signaling. In most plants, including vegetable species, intercellular signaling can be delivered by mobile regulators that traffic through a channel called plasmodesmata connecting almost all cells. A large number of transcription factors and RNAs have been discovered to move across plasmodesmata (called the symplastic way) to travel a short-range or a long-distance. This symplastic transport of signaling molecules has emerged to be an important regulation of a wide range of developmental and physiological processes. Callose deposition to plasmodesmata is a key step controlling the plasmodesmata permeability in many cell types. Here we summarize the recent progress in our understanding of plasmodesmata-mediated signaling in plants.
-
Key words:
- Intercellular /
- Signaling /
- Plasmodesmata












