-
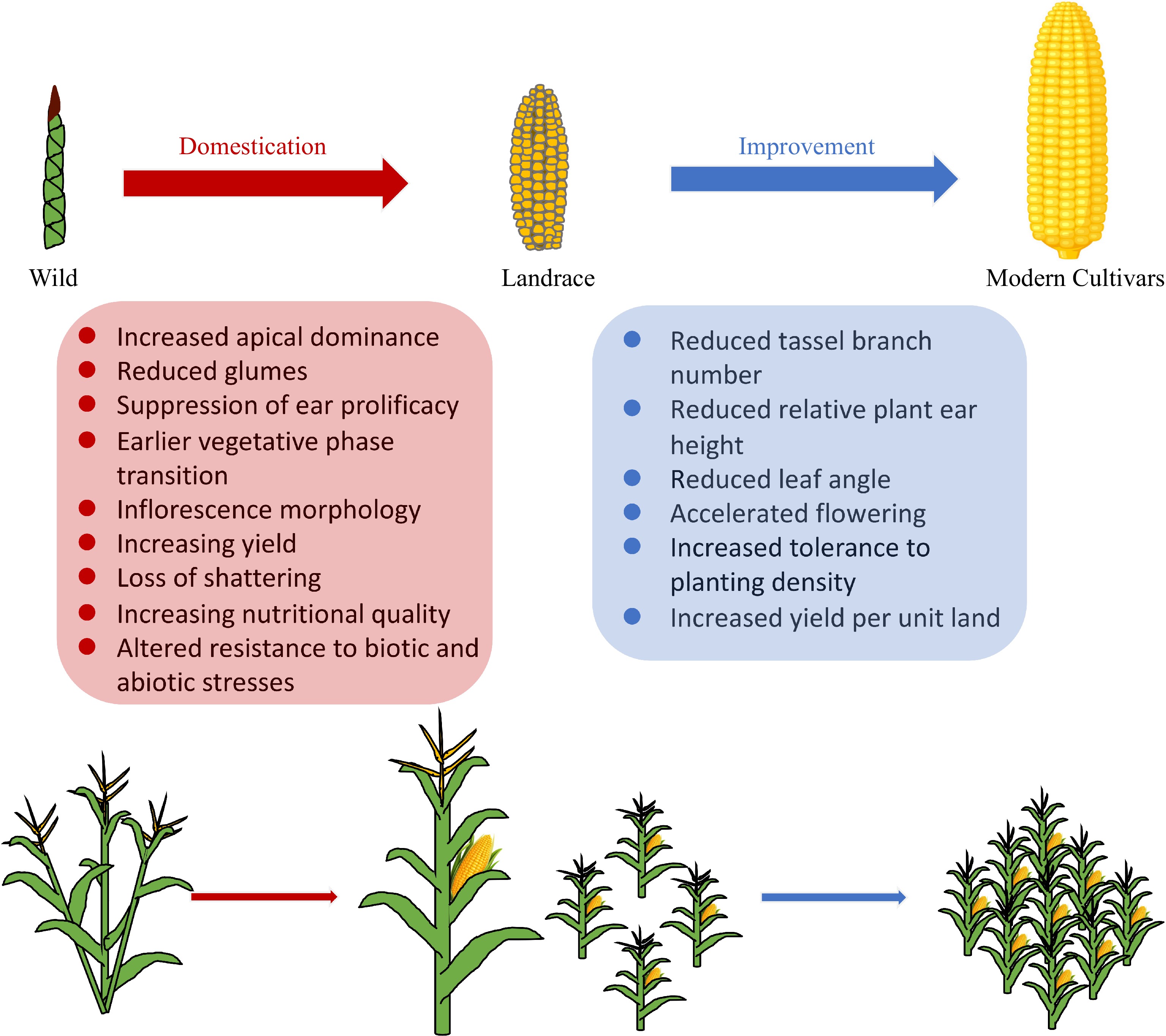
Figure 1.
Main traits of maize involved in domestication and improvement.
-
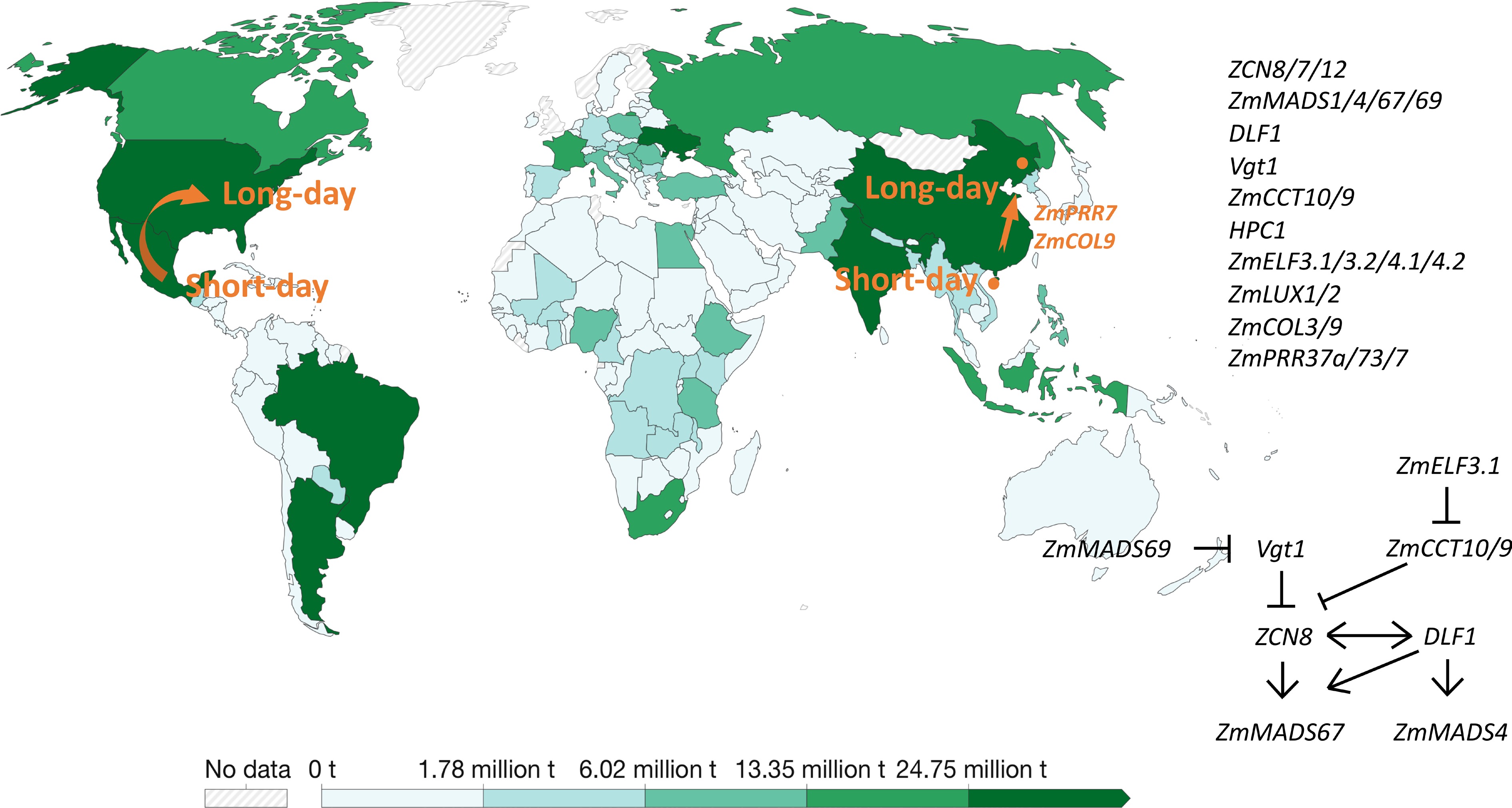
Figure 2.
Genes involved in Pre-Columbia spread of maize to higher latitudes and the temperate regions. The production of world maize in 2020 is presented by the green bar in the map from Ritchie et al. (2023). Ritchie H, Rosado P, and Roser M. 2023. "Agricultural Production". Published online at OurWorldInData.org. Retrieved from: 'https:ourowrldindata.org/agricultural-production' [online Resource].
-
Gene Phenotype Functional annotation Selection type Causative change References tb1 Plant architecture TCP transcription factor Increased expression ~60 kb upstream of tb1 enhancing expression [18−22] tga1 Hardened fruitcase SBP-domain transcription factor Protein function A SNP in exon (K-N) [25, 26] gt1 Plant architecture Homeodomain leucine zipper Increased expression prol1.1 in 2.7 kb upstream of the promoter region increasing expression [27, 28] Zm00001d020683 Plant architecture INDETERMINATE DOMAIN transcription factor Protein function Unknown [29] UPA1 Leaf angle Brassinosteroid C-6 oxidase1 Protein function Unknown [30] UPA2 Leaf angle B3 domain transcription factor Increased expression A 2 bp indel in 9.5 kb upstream of ZmRALV1 [30] Gl15 Vegetative phase change AP2-like transcription factor Altered expression SNP2154: a stop codon (G-A) [34, 35] tru1 Plant architecture BTB/POZ ankyrin repeat protein Increased expression Unknown [36] ra1 Inflorescence architecture Transcription factor Altered expression Unknown [37, 38] zfl Plant architecture Transcription factor Altered expression Unknown [40, 41] UB3 Kernel row number SBP-box transcription factor Altered expression A TE in the intergenic region; [44−46] SNP (S35): third exon of UB3
(A-G) increasing expression of UB3 and KRNKRN1/ids1/Ts6 Kernel row number AP2 Transcription factor Increased expression Unknown [47, 48] KRN2 Kernel row number WD40 domain Decreased expression Unknown [50] qHKW1 Kernel row weight CLV1/BAM-related receptor kinase-like protein Increased expression 8.9 kb insertion upstream of HKW [51, 52] ZmVPS29 Kernel morphology A retromer complex component Protein function Two SNPs (S-1830 and S-1558) in the promoter of ZmVPS29 [53] ZmSWEET4c Seed filling Hexose transporter Protein function Unknown [54] ZmSh1 Shattering A zinc finger and YABBY transcription factor Protein function Unknown [57, 58] Thp9 Nutrition quality Asparagine synthetase 4 enzyme Protein function A deletion in 10th intron of Thp9 reducing NUE and protein content [63] ZmMM1 Biotic stress MYB Transcription repressor Protein function Unknown [69] ZmHKT1 Abiotic stress A sodium transporter Protein function SNP947-G: a nonsynonymous variation increasing salt tolerance [70] ZmSRO1d-R Drought resistance and production PolyADP-ribose polymerase and C-terminal RST domain Protein function Three non-synonymous variants: SNP131 (A44G), SNP134 (V45A) and InDel433 [71] Table 1.
Key domestication genes cloned in maize.
-
Gene Functional annotation Causative change References ZCN8 Florigen protein SNP-1245 and Indel-2339 in promoter [73, 74] DLF1 Basic leucine zipper transcription factor Unknown [75] ZmMADS69 MADS-box transcription factor Unknown [76] ZmRap2.7 AP2-like transcription factor MITE TE inserted ~70 kb upstream [77−79] ZmCCT CCT-domain protein 5122-bp CACTA-like TE inserted ~2.5 kb upstream [72,81] ZmCCT9 CCT transcription factor A harbinger-like element at 57 kb upstream [82] ZmELF3.1 Unknown wo retrotransposons in the promote [84] HPC1 Phospholipase A1 enzym Unknown [83] ZmPRR7 Unknown Unknown [88] ZmCOL9 CO-like-transcription factor Unknown [88] Table 2.
Flowering time related genes contributing to Pre-Columbia spread of maize.
-
Gene Phenotype Functional annotation Selection type Causative change References ZmPIF3.1 Plant height Basic helix-loop-helix transcription factor Increased expression Unknown [91] TSH4 Tassel branch number Transcription factor Altered expression Unknown [91] ZmPGP1 Plant architecture ATP binding cassette transporter Altered expression A 241 bp deletion in the last exon of ZmPGP1 [92, 93] PhyB2 Light signal Phytochrome B Altered expression A 10 bp deletion in the translation start site [101] ZmPIF4.1 Light signal Basic helix-loop-helix transcription factor Altered expression Unknown [102] ZmKOB1 Grain yield Glycotransferase-like protein Protein function Unknown [121] Table 3.
Selective genes underpinning genetic improvement during modern maize breeding.
Figures
(2)
Tables
(3)