-
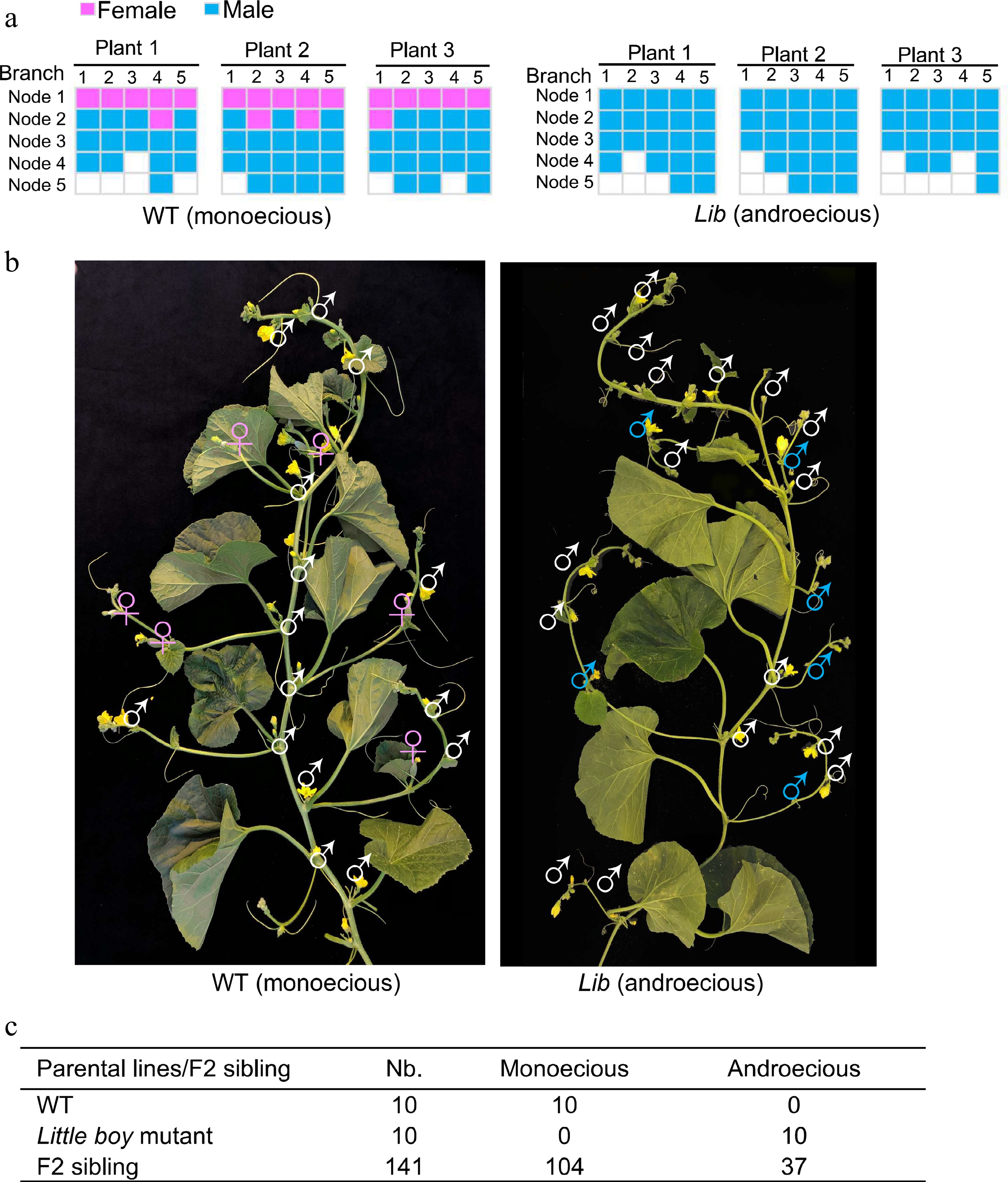
Figure 1.
Sexual morphs observed in monoecious melon and androecious little boy mutant[7]. (a) Graphical presentation of flower sexual types observed in wild type monoecious melon (Cucumis melo) and Lib (Little boy) androecious mutant. (b) Phenotype of Lib mutant. Female flowers (in pink) were transformed into male flowers (in blue) in Lib mutant. The inflorescence structure in the Lib mutant is not affected. (c) Segregation analysis of androecious phenotype in F2 populations. Nb, total number of plants.
-
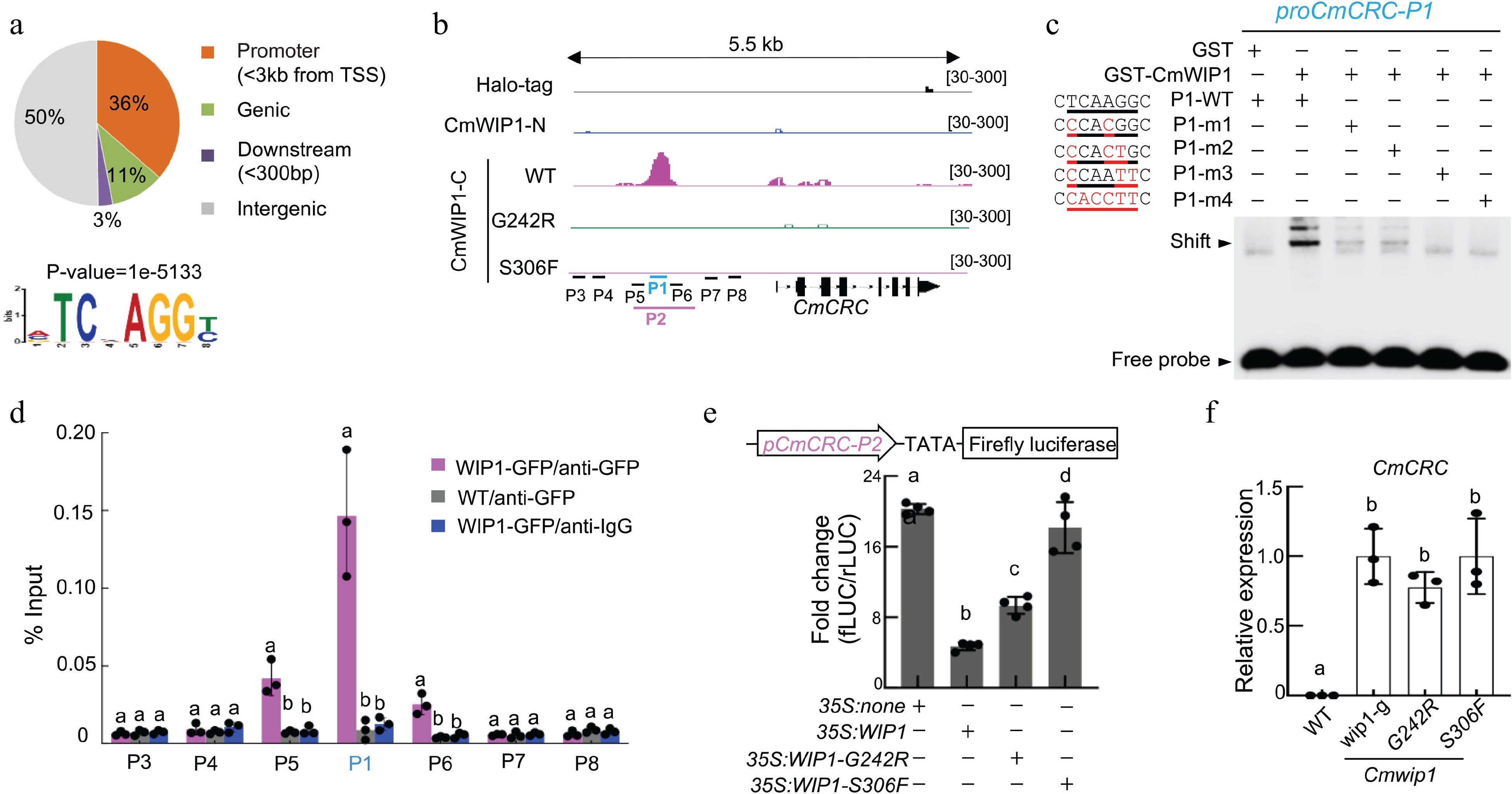
Figure 2.
CmWIP1 binds to the CRC promoter[7]. (a) Distribution of CmWIP1 DAP-seq peaks and CmWIP1 binding motif. (b) CmWIP1 binding at the CmCRC locus. Halo-tag, negative control; -N, N terminus; -C, C terminus; P1 to P8, promoter sequences used in EMSA, ChIP, and promoter activity assays. G242R and S306F mutations in WIP1 DNA binding domain. (c) EMSA assay. A5-μM GST or WIP1-GST protein and a 10-nM DNA probe were used. P1-m1 to P1-m4, mutant probes. GST, Glutathione S-transferase. (d) ChIP-qPCR assay in melon protoplast transformed with 35S:WIP1-GFP. Results are expressed as a percentage of the INPUT fraction. Wild-type protoplasts and anti-IgG antibody were used as negative controls. (e) Promoter activity assays. Empty vector (35S: none), 35S:WIP1 or variants were co-transformed into melon protoplasts along with pCmCRC-P2: fLUC. Bars represent means ± SD (n = 4). (f) qRT-PCR analysis of CmCRC. Flower buds at stage 6 were used. wip1 (g), a natural epigenetic mutant; G242R and S306F, TILLING mutants (14). In (d)−(f), bars represent mean ± SD (n = 3) and different letters denote significant differences (p < 0.05, one-way ANOVA with Tukey's post hoc test).
-
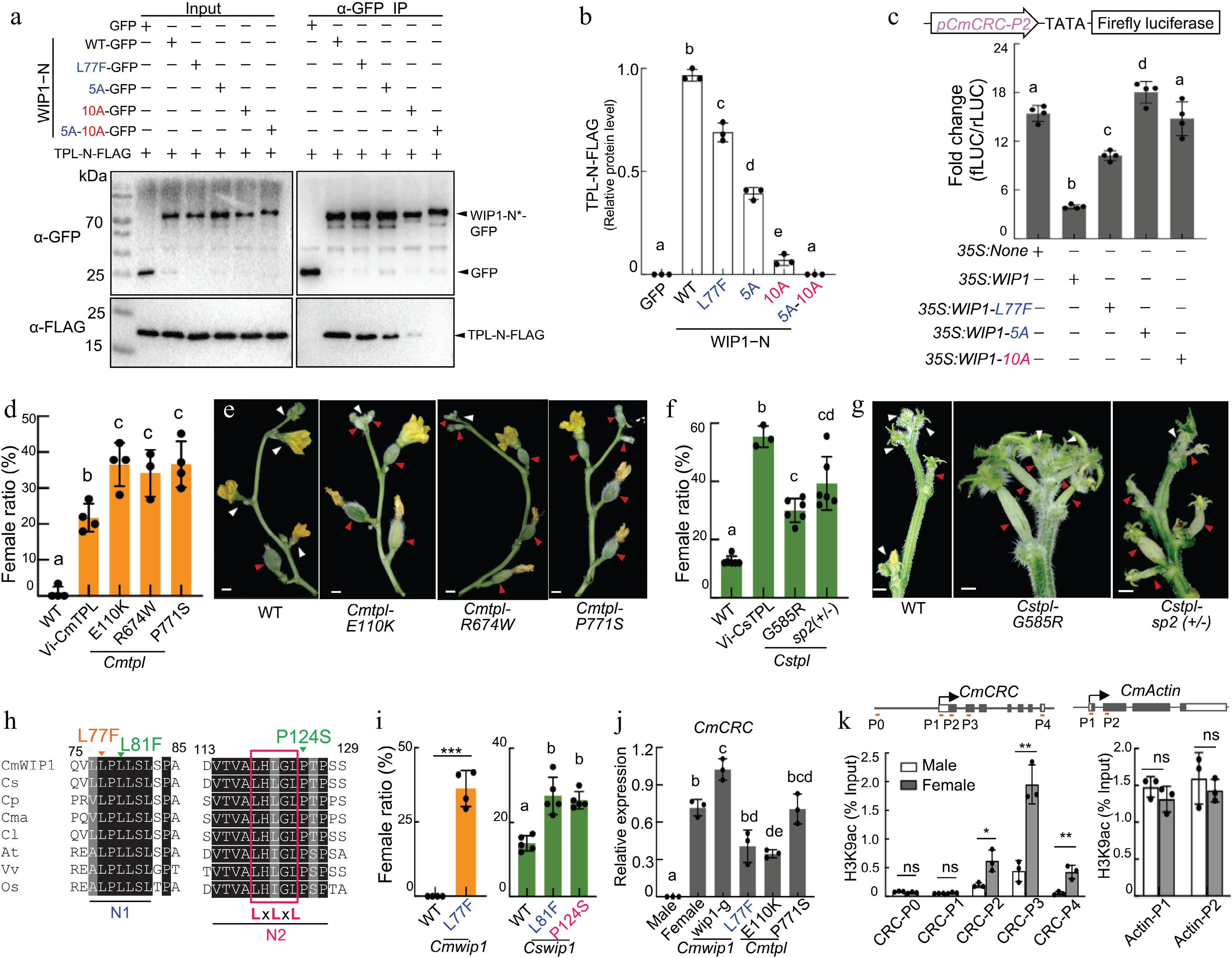
Figure 3.
WIP1 recruits Topless to repress CRC[7]. (a) Co-IP assays. GFP-tagged WIP1−N or variants (asterisk) were co-transformed into melon protoplasts along with FLAG-tagged TPL-N. (b) Quantification of FLAG-tagged TPL-N from Co-IP assays in (a). (c) Promoter activity assay. Empty vector (35S: none), 35S:WIP1 or variants were co-transformed into melon protoplasts along with pCmCRC-P2: fLUC. Statistical analysis of female flower ratio in monoecious (WT), VIGS-CmTPL (Vi-CmTPL) and tpl mutants in (d) melon and (f) cucumber. Flower phenotypes of tpl mutants in (e) melon and (g) cucumber; white and red triangles denote male and female flowers, respectively; scale bars, 5 mm. (h) Sequence alignment of WIP1 and homologous proteins showing N1 and N2 motifs. Initials of species are indicated. LxLxL, EAR motif. Induced mutations in melon (orange) and cucumber (green) are shown. (i) The female ratio [females/(females + males)], in percent, of nodes 2 to 10 of lateral branches (melon) and nodes 1 to 20 of main stem (cucumber). qRT-PCR analysis of (j) CmCRC and ChIP-qPCR analysis of (k) H3K9ac at stage 6 flower buds. CmACTIN1, control. Bars represent means ± SD (n = 3−5); different letters denote significant differences p < 0.05, one-way ANOVA with Tukey’s post hoc test). * p < 0.05, ** p < 0.01 (Student’s t test).
-
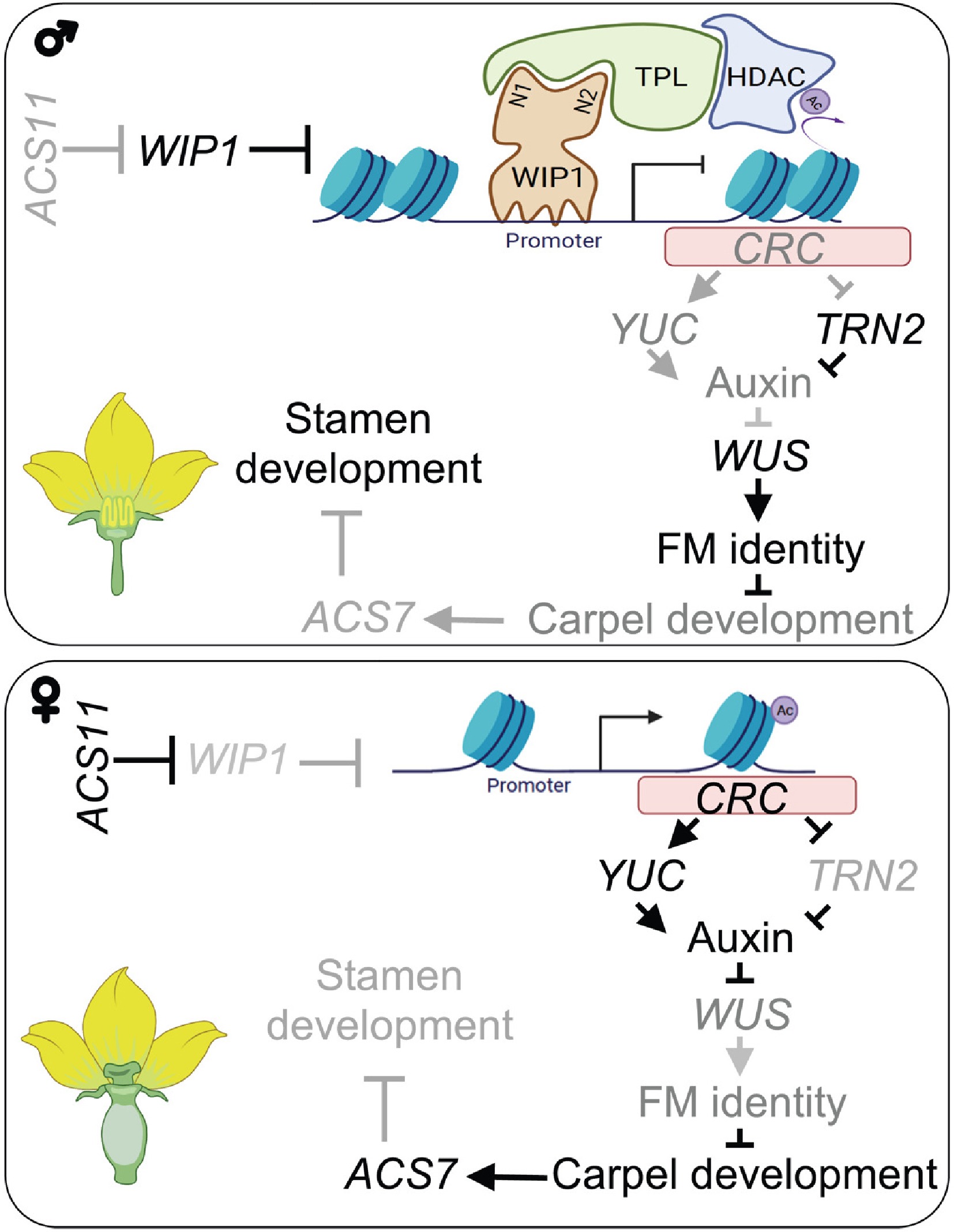
Figure 4.
Model explaining how WIP1-TPL controls the expression of CRC to lead to male flower development[7].
Figures
(4)
Tables
(0)