-
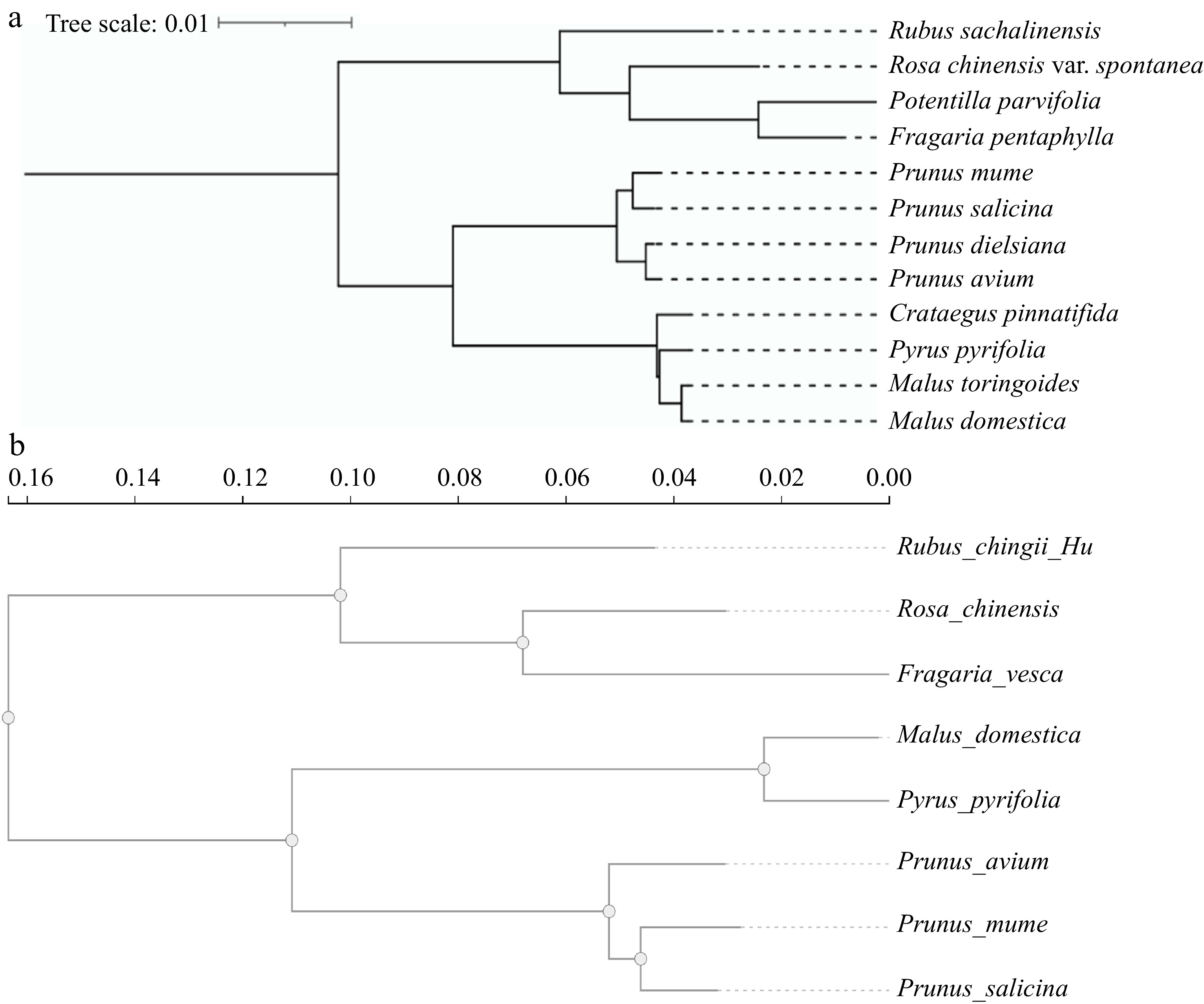
Figure 1.
Phylogenetic evolution tree of Rosaceae plants. (a) The Rosaceae represents the plant chloroplast genome evolutionary tree. (b) The Rosaceae represents the plant nuclear genome evolutionary tree.
-
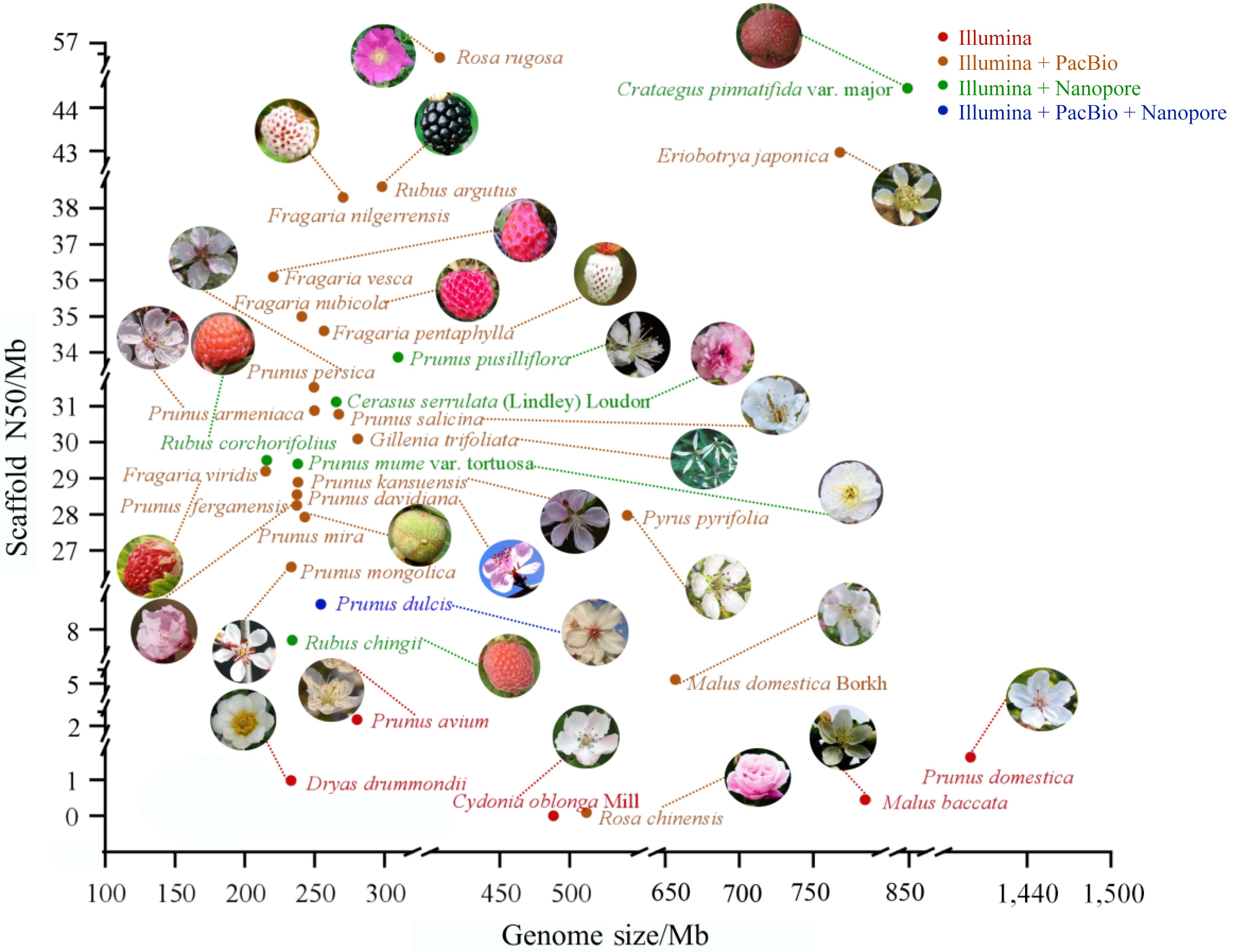
Figure 2.
Summary of plant genome sequencing for Rosaceae. Each plant's genome size is shown by the x-axis, and the scaffold N50 of the genome assembly is represented by the y-axis. Different colors correspond to different sequencing platforms.
-
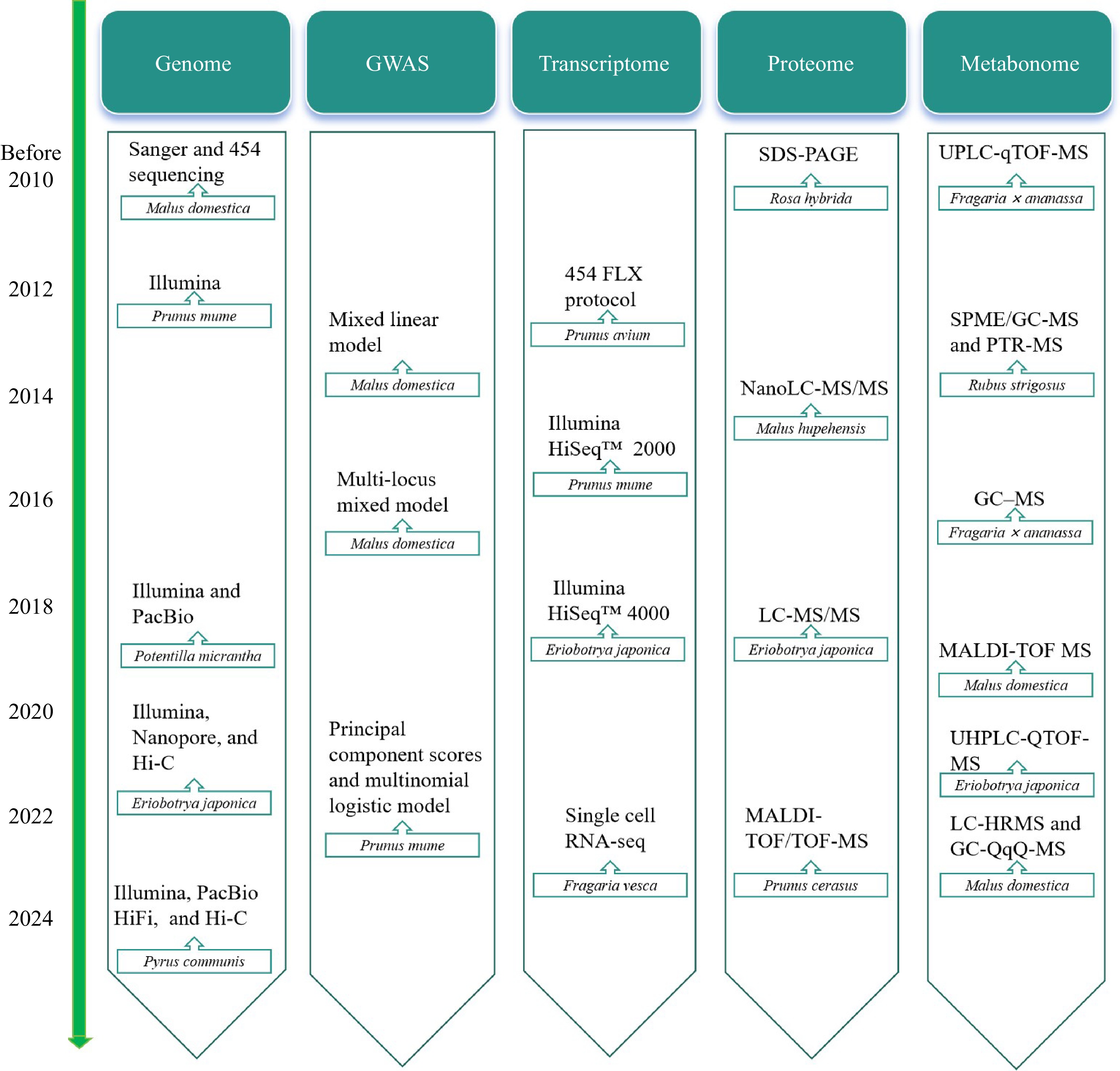
Figure 3.
Timeline for omics research on Rosaceae plants.
-
Species Sequencing method Genome size (Mb) Scaffold N50 (Mb) Ref. Cerasus serrulata (Lindley) Loudon Illumina + Nanopore 265.40 31.12 [46] Prunus avium Illumina 280.33 2.48 [47] Prunus pusilliflora Illumina + Nanopore 309.62 33.87 [48] Dryas drummondii Illumina 233.00 0.98 [49] Eriobotrya japonica Illumina + Nanopore 760.98 43.16 [50] Gillenia trifoliata PacBio 280.76 30.09 [50] Fragaria vesca Illumina + PacBio 220.40 36.1 [51] Fragaria nilgerrensis Illumina + PacBio 270.30 38.3 [52] Fragaria viridis Illumina + PacBio 214.90 29.2 [53] Fragaria nubicola Illumina + PacBio 240.75 35 [53] Fragaria pentaphylla Illumina + PacBio 256.74 34.6 [54] Prunus armeniaca Illumina + PacBio 249.73 30.88 [37] Prunus persica Illumina + PacBio 249.41 31.53 [37] Prunus davidiana Illumina + PacBio 237.29 28.55 [55] Prunus kansuensis Illumina + PacBio 238.06 28.89 [55] Prunus ferganensis Illumina + PacBio 237.24 28.25 [55] Prunus mume var. tortuosa Illumina + Nanopore 237.80 29.4 [56] Pyrus pyrifolia Illumina + PacBio 541.30 27.97 [44] Prunus salicina Illumina + PacBio 267.18 30.78 [37] Prunus domestica Illumina 1,399.32 1.63 [57] Malus domestica Borkh. Illumina + PacBio 643.2 5.5 [58] Malus baccata Illumina 779.00 0.45 [59] Prunus mira Illumina + PacBio 242.94 27.93 [55] Prunus dulcis Illumina + PacBio + Nanopore 254.45 9.2 [60] Prunus mongolica Illumina + PacBio 233.17 26.54 [61] Cydonia oblonga Mill. Illumina 488.40 0.0024 [62] Crataegus pinnatifida var. major Illumina + Nanopore 856.88 44.94 [63] Rosa chinensis Illumina + PacBio 512.00 0.09 [64] Rosa rugosa Illumina + PacBio 407.00 56.6 [65] Rubus chingii Hu Illumina + Nanopore 219.80 8.2 [66] Rubus corchorifolius Illumina + Nanopore 215.74 29.5 [67] Rubus argutus Illumina + PacBio 298.24 38.6 [68] Table 1.
Progression genome sequencing of Rosaceae.
-
Omics analysis Application level Species Omics analysis Application level Species Comparative genomics Species evolution Rubus corchorifolius GWAS Fusarium wil Fragaria × ananassa Prunus davidiana,
Prunus mira,
Prunus kansuensis,
Prunus ferganensisFlesh browning Malus domestica Prunus pusilliflora Salt and drought stress Rosa chinensis Transcriptome Salt stress Prunus mume Cold stress Prunus mume Heat stress Rosa chinensis Flower color Prunus mume Drought stress Rosa chinensis Prunus persica f. versicolor Botrytis. cinerea Rosa hybrida Prunus campanulata 'Plean' Fragaria vesca Fruit morphology Prunus persica, Prunus salicina, Prunus armeniaca, Prunus mira, Prunus davidiana Cold stress Rosa hybrida Fruit aroma Malus domestica Eriobotrya japonica Prunus persica Rosa xanthina f. spontanea Fruit flavor Pyrus spp Prunus pyrifolia Metabolomics Drought and salt stress Fragaria × ananassa Flower color Pink-flowered strawberry Cold stress Prunus mume Malus halliana Malus domestica Rosa chinensis
Rosa rugosaNitrogen and phosphorus absorption Malus domestica Flower color Miniature rose Fruit color Fragaria × ananassa Rosa hybrida Malus domestica Floral fragrance Rosa rugosa Prunus avium Prunus mume Crataegus pinnatifida Fruit flavor Eriobotrya japonica Fruit flavor Malus domestica Fragaria × ananassa Proteomics Abiotic stress Rosa hybrida Malus domestica Cold stress Fragaria × ananassa Fruit color Fragaria × ananassa Rosa beggeriana Active metabolites Rubus idaeus Salt stress Prunus cerasus Eriobotrya japonica Ring rot Malus domestica Malus domestica Flower color Prunus persica Multi-omics Salt stress Malus halliana Rosa hybrida Heat stress Eriobotrya japonica Prunus persica f. versicolor Nitrogen absorption Malus domestica Prunus mume Weeping trait Prunus mume Fruit development Malus halliana Flower development Eriobotrya japonica Fruit storage Prunus persica Floral fragrance Prunus mume Fruit aroma Prunus persica Fruit flavor Malus domestica Fruit color Cerasus humilis Active metabolites Eriobotrya japonica Table 2.
The main findings of omics analysis of Rosaceae plants.
Figures
(3)
Tables
(2)