-
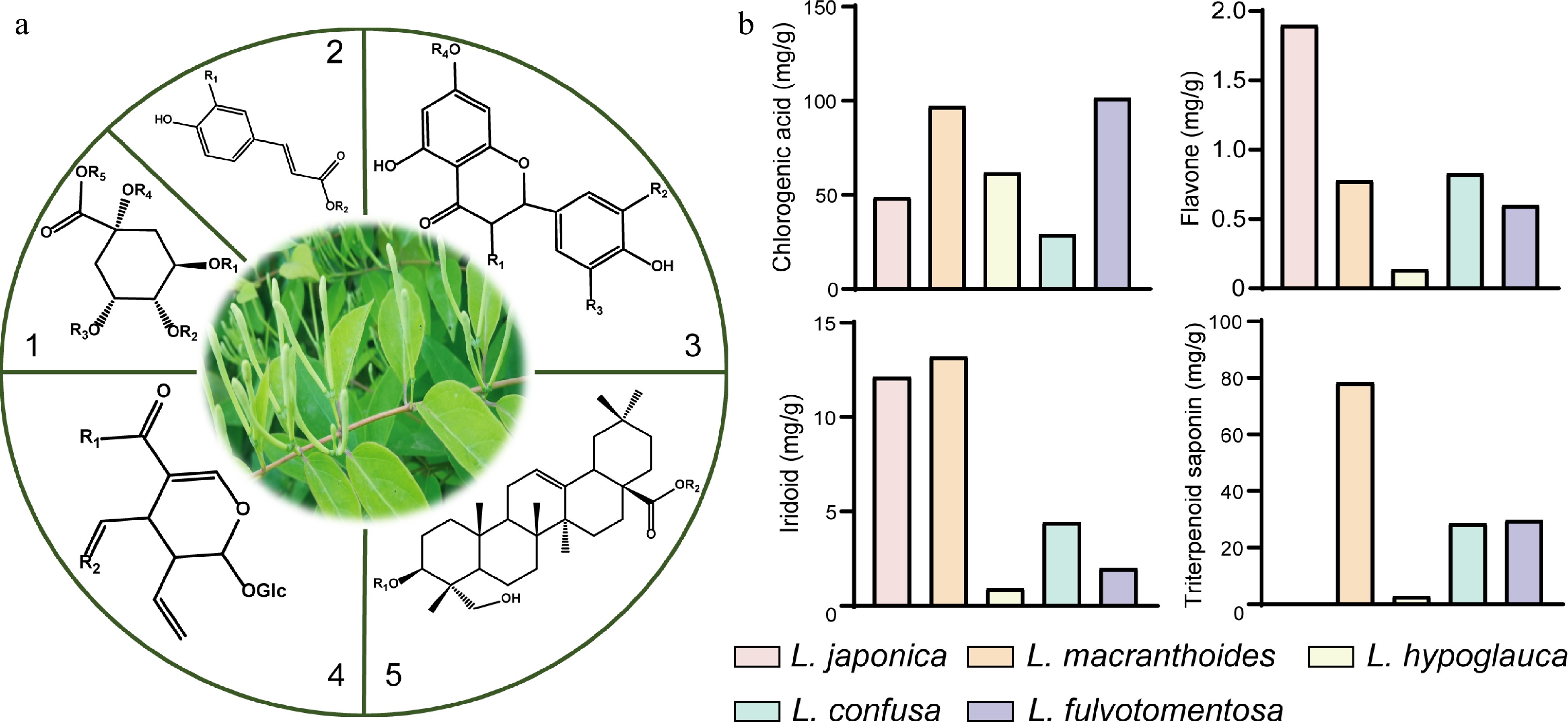
Figure 1.
Core structures of main secondary metabolites and their distribution in five species of Lonicera. (a) 1 and 2, the main core structures of organic acids; 3, the main core structures of flavonoids; 4, the main core structures of iridoids; 5, the main core structures of triterpene saponins. (b) Comparison of dry weight of four kinds of substances in five species of Lonicera[17,28].
-
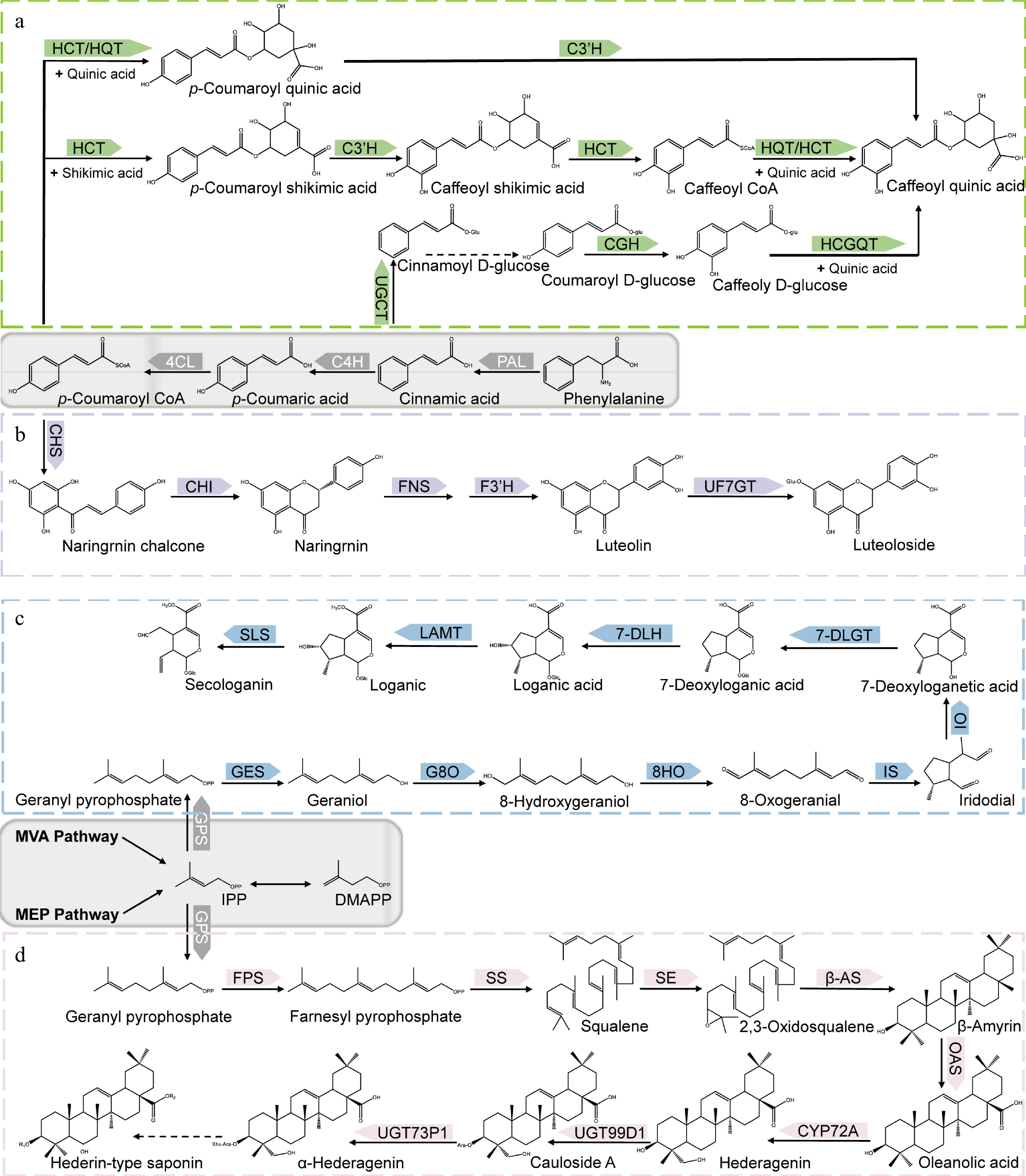
Figure 2.
Biosynthetic pathways of main bioactive constituents of Lonicera. (a) Biosynthetic pathways of chlorogenic acid. (b) Biosynthetic pathways of luteoloside. (c) Biosynthetic pathways of secologanin. (d) Biosynthetic pathways of hederin-type triterpene saponins. PAL, phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-hydroxycinnamoyl CoA ligase; HCT, hydroxycinnamoyl CoA shikimate/quinate hydroxycinnamoyl transferase; C3’H, p-coumaroyl 3-hydroxylase; HQT, hydroxycinnamoyl-CoA quinate transferase; UGCT, UDP glucose: cinnamate glucosyl transferase; CGH, p-coumaroyl-D-glucose hydroxylase; HCGQT, hydroxycinnamoyl D-glucose: quinate hydroxycinnamoyl transferase; CHS, Chalcone synthase; CHI, Chalcone isomerase; FNS, Flavone synthase; F3H, flavonoid 30-monooxygenase/flavonoid 30-hydroxylase; UF7GT, flavone 7-O-β-glucosyltransferase; GPS, Geranyl pyrophosphatase; GES, geraniol synthase; G8O, geraniol 10-hydroxylase/8-oxidase; 8HO, 8-hydroxygeraniol oxidoreductase; IS, iridoid synthase; IO, iridoid oxidase; 7DLGT, 7-deoxyloganetic acid glucosyltransferase; 7DLH, 7-deoxyloganic acid hydroxylase; LAMT, loganic acid O-methyltransferase; SLS, secologanin synthase; FPS, farnesyl pyrophosphate synthase; SS, squalene synthase; SE, squalene epoxidase; β-AS, β-amyrin synthase; OAS, oleanolic acid synthetase.
-
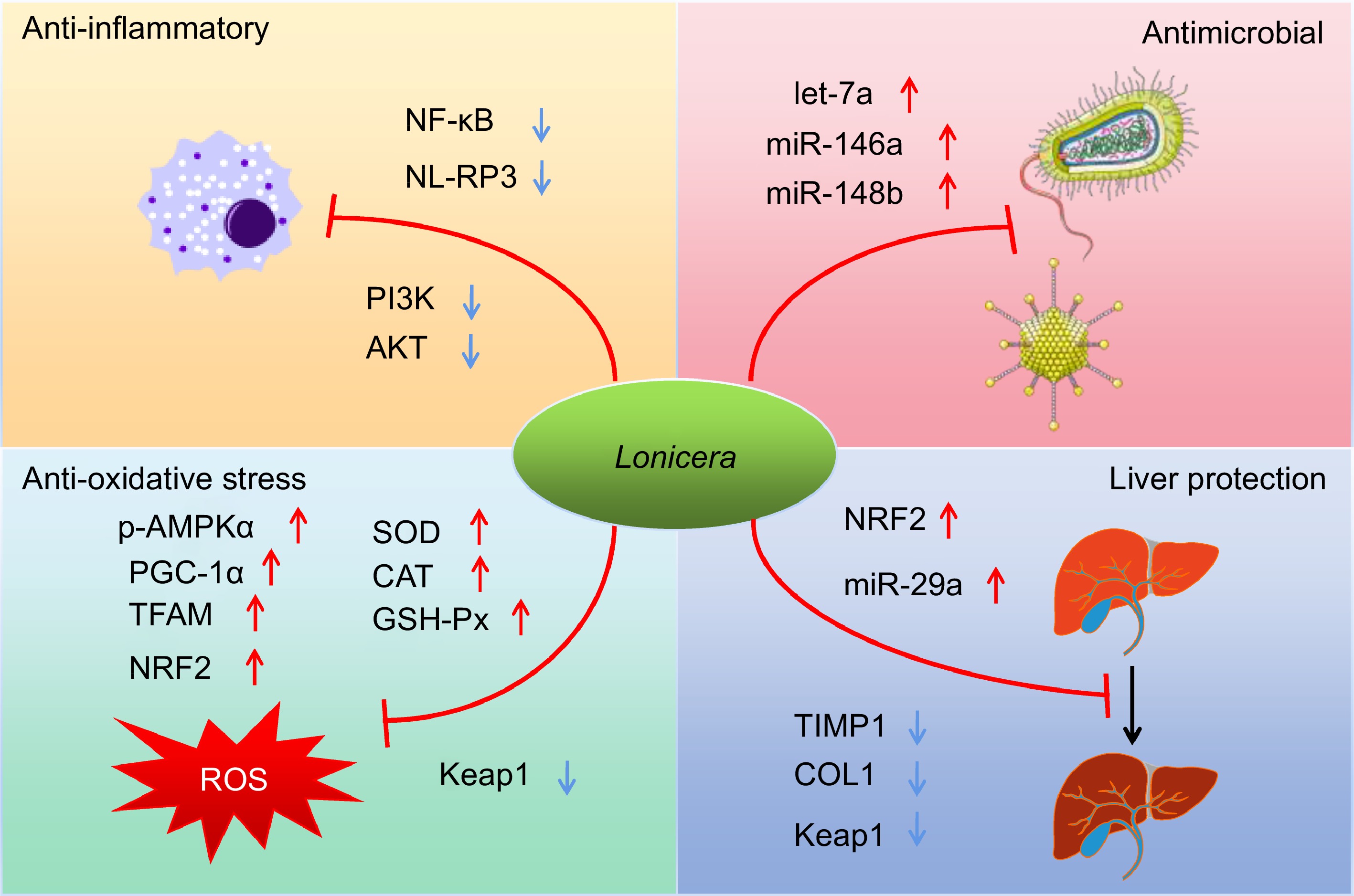
Figure 3.
Schematic summary of four main pharmacological effects (anti-inflammatory, antimicrobial, anti-oxidative and hepatoprotective effects) of the Lonicera genus and the underlying mechanisms of actions.
-
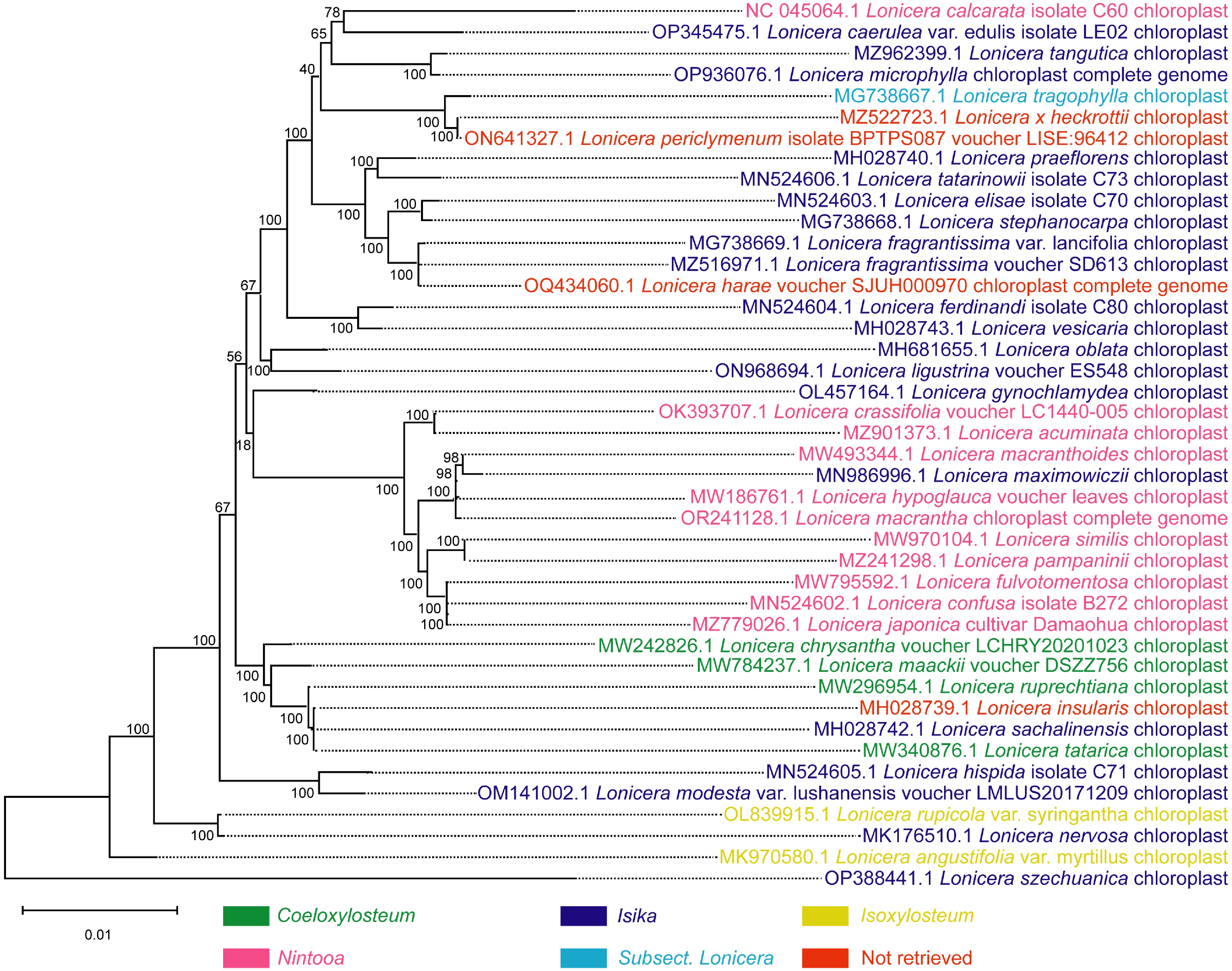
Figure 4.
Phylogenetic tree of 42 species of the Lonicera genus based on complete chloroplast genome sequence data. The phylogenetic tree was constructed by the maximum likelihood method. Coeloxylosteum, Isika, Isoxylosteum, and Nintooa belong to Chamaecerasus and Subsect. Lonicera belongs to the Periclymenum. Chamaecerasus and Periclymenum are the two subgenera of Lonicera. 'Not retrieved' indicates that the species failed to retrieve a subordinate taxon in the Lonicera.
-
Engineering bacteria Operational methods Products Yield Refs S. cerevisiae Eliminate the tyrosine-induced feedback inhibition, delete genes involved in competing pathways and overexpress rate-limiting enzymes Caffeic acid 569.0 mg/L [69] S. cerevisiae Employe a heterologous tyrosine ammonia lyase and a 4HPA3H complex composed of HpaB and HpaC derived from different species Caffeic acid 289.4 mg/L [73] S. cerevisiae Supply and recycle of three cofactors: FADH2, S-adenosyl-L-methion, NADPH Caffeic acid
Ferulic acidCaffeic acid: 5.5 g/L;
Ferulic acid: 3.8 g/L[117] E. coli Knocking out competing pathways Caffeic acid 7,922 mg/L [118] E. coli Artificial microbial community, a polyculture of three recombinant Escherichia coli strains Chlorogenic acid 250 μM [68] Cell-free biosynthesis Extract and purify spy-cyclized enzymes (CFBS-mixture) Chlorogenic acid 711.26 mg/L [70] S. cerevisiae Three metabolic engineering modules were systematically optimized: shikimate pathway and carbon distribution, branch pathways, CGA pathway genes Chlorogenic acid Flask fermentation: 234.8 mg/L;
Fed-batch fermentation:
806.8 mg/L[119] E. coli Using modular coculture engineering: construction of the defective strain improves the production and utilization of precursor substances Chlorogenic acid 131.31 mg/L [122] E. coli Introduce heterologous UDP-glucose biosynthetic genes Luteolin 34 mg/L [120] Y. lipolytica Overexpression of the key genes involved in the mevalonate pathway, the gene encoding cytochrome P450 (CYP716A12) to that encoding NADPH-P450 reductase Oleanolic acid 129.9 mg/L [85] S. cerevisiae Improve the pairing efficiency between Cytochrome P450 monooxygenase and reductase and the expression level of key genes Oleanolic acid 606.9 mg/L [121] S. cerevisiae Heterologous expression and optimization of CrAS, CrAO, and AtCPR1, and regulation of ERG1 and NADPH regeneration system Oleanolic acid 433.9 mg/L [123] Table 1.
Biosynthesis of Lonicera-specialized metabolites using metabolic engineering.
Figures
(4)
Tables
(1)