-
The Lonicera Linn. genus is a constituent member of the Caprifoliaceae family[1]. It is the largest genus in this family and comprises at least 200 species with a notable presence in North Africa, North America, Asia, and Europe[1]. Members of the Lonicera genus possess a wide range of economic benefits from their use as ornamental plants to food and as plants credited with numerous health benefits. Conspicuous among the numerous members of this genus with known medicinal uses are L. japonica, L. macranthoides, L. hypoglauca, L. confusa, and L. fulvotomentosa[2]. Though these species feature prominently in the Chinese Pharmacopoeia, other species such as L. acuminata, L. buchananii, and L. similis are recognized as medicinal resources in certain parts of China[1]. Among the aforementioned species, L. japonica takes precedence over the rest due to its high medicinal and nutritional value[3,4]. For instance, the microRNA MIR2911, an isolate from L. japonica, has been reported to inhibit the replication of viruses[5−7]. Also, the water extract of L. japonica has been used to produce various beverages and health products[8]. The Lonicera genus therefore possesses huge prospects in the pharmaceutical, food, and cosmetic industries as an invaluable raw material[9].
The main active constituents of the Lonicera genus include organic acids, flavonoids, iridoids, and triterpene saponins. Chlorogenic acids, iridoids, and flavones are mainly credited with the anti-inflammatory, antiviral, anticancer, and antioxidant effects of the various Lonicera species[10−13]. Their hepatoprotective, immune modulatory, anti-tumor and anti-Alzheimer’s effects are for the most part ascribed to the triterpene saponins[14−16]. As stated in the Chinese Pharmacopoeia and backed by the findings of diverse research groups, the plants of the Lonicera genus are known to possess high amounts of organic acids (specifically chlorogenic acid) and pentacyclic triterpenoid saponins[2,17−19]. The flower and flower bud have traditionally served as the main medicinal parts of the Lonicera genus even though there is ample evidence that the leaves possess the same chemical composition[20]. A perusal of the current scientific literature reveals the fact that little attention has been devoted to exploring the biosynthesis of the chemical constituents of the Lonicera genus with the view to finding alternative means of obtaining higher yields. It is therefore imperative that priority is given to the exploration of the biosynthesis of these bioactive compounds as a possible means of resource protection. There is also the need for further research on ways to fully tap the medicinal benefits of other plant parts in the Lonicera genus.
Here, we provide a comprehensive review of relevant scientific literature covering the structure, pharmacology, multi-omics analyses, phylogenetic analysis, biosynthesis, and metabolic engineering of the main bioactive constituents of the Lonicera genus. Finally, we proffer suggestions on the prospects of fully exploiting and utilizing plants of the Lonicera genus as useful medicinal plant resources.
-
A total of at least 400 secondary metabolites have been reported for the Lonicera genus. These metabolites are categorized into four main groups (Fig. 1a), including not less than 50 organic acids, 80 flavonoids, 80 iridoids, and 80 triterpene saponins[21−23]. Organic acids are mainly derivatives of p-hydroxycinnamic acid and quinic acid. Among the organic acids, chlorogenic acids are reported to be the main bioactive compounds in L. japonica[24−26]. The organic acids are most abundant in the leaves, while the least amounts are found in the stem of L. japonica. The flowers of the plant are known to contain moderately high amounts of organic acids[27]. The basic core structure of the flavonoids is 2-phenylchromogen. Luteolin and its glycoside which are characteristic flavonoids of the Lonicera genus are most abundant in L. japonica[28]. On the whole, the flavonoid contents in L. japonica are also highest in the leaves, available in moderate amounts in the flowers, and in lowest amounts in the stem[21]. The core structures of the iridoids are iridoid alcohols, the chemical properties of which are similar to hemiacetal. The iridoids often exist in the form of iridoid glycosides in plants. Secoiridoids glycosides are predominant in the Lonicera genus[25]. In L. japonica, the contents of the iridoids are most abundant in the flowers, moderate in leaves, and lowest in the stem[21]. The characteristic saponins of the Lonicera genus are mainly pentacyclic triterpenoids, including the hederin-, oleanane-, lupane-, ursulane- and fernane-types, etc[22]. The hederin-type saponins are reported in the highest amounts in L. macranthoides[17] (Fig. 1b).
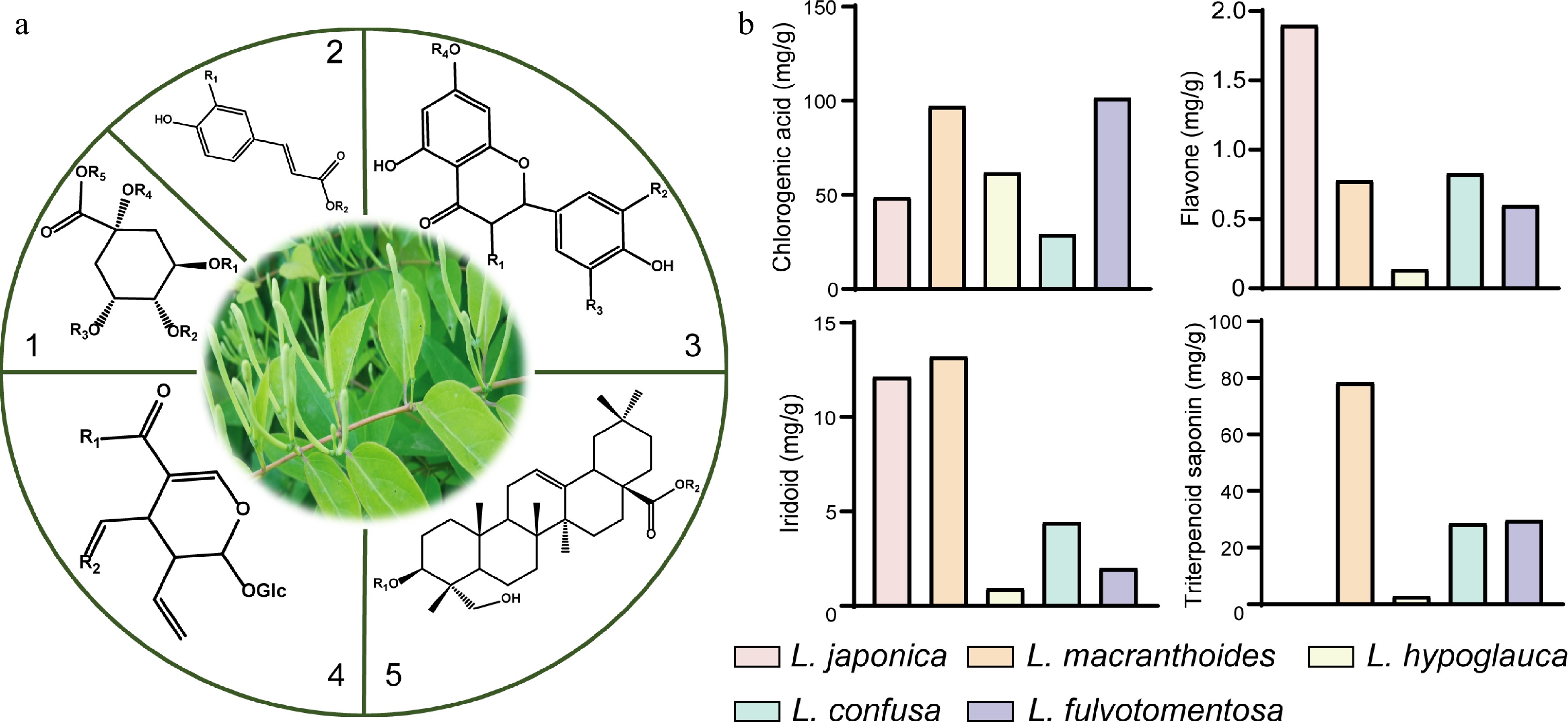
Figure 1.
Core structures of main secondary metabolites and their distribution in five species of Lonicera. (a) 1 and 2, the main core structures of organic acids; 3, the main core structures of flavonoids; 4, the main core structures of iridoids; 5, the main core structures of triterpene saponins. (b) Comparison of dry weight of four kinds of substances in five species of Lonicera[17,28].
-
The similarities between chlorogenic acid (CGA) and flavonoids can be traced back to their biosynthesis since p-coumaroyl CoA serves as the common precursor for these compounds[29]. p-coumaroyl CoA is obtained through sequential catalysis of phenylalanine and its biosynthetic intermediates by phenylalanine-ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H) and 4-coumarate CoA ligase (4CL)[30−33].
CGA is a phenolic acid composed of caffeic acid and quinic acid and is the most important bioactive compound among the organic acids. Its biosynthesis has been relatively well-established; three main biosynthetic routes have been propounded (Fig. 2a). One route relates to the catalysis of caffeoyl-CoA and quinic acid by hydroxycinnamoyl-CoA quinate transferase (HQT)/hydroxycinnamoyl CoA shikimate/quinate hydroxycinnamoyl transferase (HCT) to produce CGA[34−37]. The HQT-mediated pathway has been deemed the major route for CGA synthesis in in different plant species[38,39]. The second biosynthetic route stems from the biosynthesis of p-coumaroyl quinate through the catalytic effect of HCT/HQT and subsequent hydroxylation of p-coumaroyl quinate under the catalysis of p-coumarate 3'-hydroxylase (C3’H)[34,36,37]. For the third route, caffeoyl glucoside serves as the intermediate to form CGA, a process that is catalyzed by hydroxycinnamyl D-glucose: quinic acid hydroxycinnamyl transferase (HCGQT)[40,41].
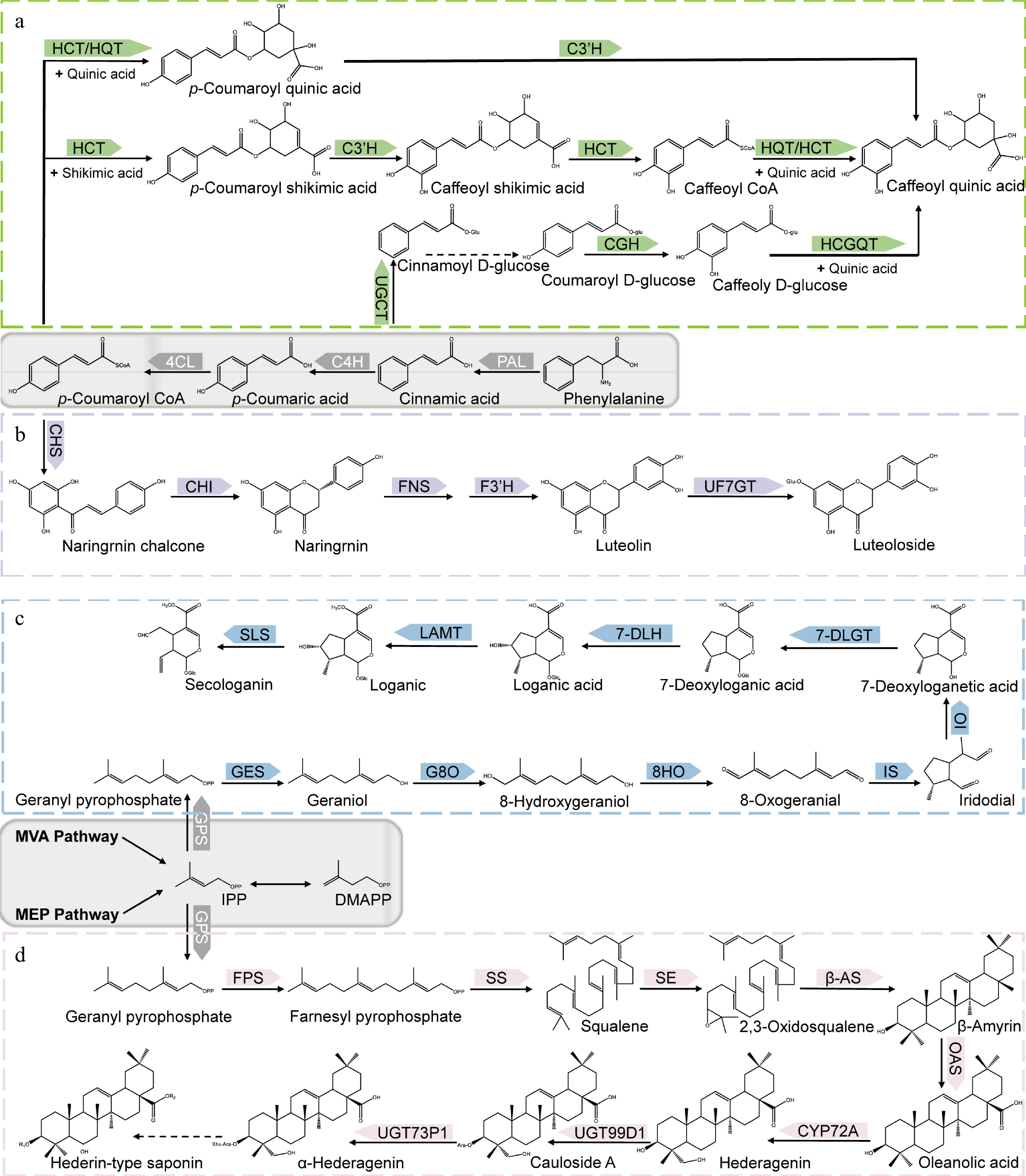
Figure 2.
Biosynthetic pathways of main bioactive constituents of Lonicera. (a) Biosynthetic pathways of chlorogenic acid. (b) Biosynthetic pathways of luteoloside. (c) Biosynthetic pathways of secologanin. (d) Biosynthetic pathways of hederin-type triterpene saponins. PAL, phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-hydroxycinnamoyl CoA ligase; HCT, hydroxycinnamoyl CoA shikimate/quinate hydroxycinnamoyl transferase; C3’H, p-coumaroyl 3-hydroxylase; HQT, hydroxycinnamoyl-CoA quinate transferase; UGCT, UDP glucose: cinnamate glucosyl transferase; CGH, p-coumaroyl-D-glucose hydroxylase; HCGQT, hydroxycinnamoyl D-glucose: quinate hydroxycinnamoyl transferase; CHS, Chalcone synthase; CHI, Chalcone isomerase; FNS, Flavone synthase; F3H, flavonoid 30-monooxygenase/flavonoid 30-hydroxylase; UF7GT, flavone 7-O-β-glucosyltransferase; GPS, Geranyl pyrophosphatase; GES, geraniol synthase; G8O, geraniol 10-hydroxylase/8-oxidase; 8HO, 8-hydroxygeraniol oxidoreductase; IS, iridoid synthase; IO, iridoid oxidase; 7DLGT, 7-deoxyloganetic acid glucosyltransferase; 7DLH, 7-deoxyloganic acid hydroxylase; LAMT, loganic acid O-methyltransferase; SLS, secologanin synthase; FPS, farnesyl pyrophosphate synthase; SS, squalene synthase; SE, squalene epoxidase; β-AS, β-amyrin synthase; OAS, oleanolic acid synthetase.
The key enzymes in the biosynthesis of p-coumaroyl CoA, and invariably CGA, thus, PAL, C4H, and 4CL have been established in diverse studies such as enzyme gene overexpression/knockdown[42], enzyme activity analysis[33] and transcriptomics[18]. However, the centrality of HQT in the biosynthesis of CGA remains disputable. While some studies have reported a strong correlation between HQT expression level with CGA content and distribution[18,35,39,43,44], others found no such link[45], bringing into question the role of HQT as a key enzyme in CGA biosynthesis.
Few studies have been conducted on the regulation of CGA biosynthesis in the Lonicera genus. It was found that overexpression of the transcription factor, LmMYB15 in Nicotiana benthamiana can promote CGA accumulation by directly activating 4CL or indirectly binding to MYB3 and MYB4 promoters[46]. LjbZIP8 can specifically bind to PAL2 and act as a transcriptional repressor to reduce PAL2 expression levels and CGA content[47]. Under NaCl stress, increased PAL expression promoted the accumulation of phenolic substances in leaves without oxidative damage, a condition that was conducive to the accumulation of the bioactive compounds in leaves[48].
Luteolin and its derivative luteolin 7-O- glucoside (luteoloside) are representative flavonoids of the Lonicera genus. Similar to CGA, luteolin is biosynthesized from p-coumaroyl CoA but via a different route. The transition from p-coumaroyl CoA to luteolin is underpinned by sequential catalysis by chalcone synthetase (CHS), chalcone isomerase (CHI), flavonoid synthetase (FNS), and flavonoid 3'-monooxygenase/flavonoid 3'-hydroxylase (F3'H)[45,49,50] (Fig. 2b). Luteoloside is synthesized from luteolin by UDP glucose-flavonoid 7-O-β-glucosyltransferase (UF7GT)[51]. Similar to CGA biosynthesis, the key enzymes of luteolin synthesis include PAL, C4H, and 4CL in addition to FNS[33,45,52]. The content of luteoloside was found to be highly abundant in senescing leaves relative to other tissues such as stem, flowers, and even young leaves[52]. Through transcriptomic analysis, luteoloside biosynthesis-related differentially expressed unigenes (DEGs) such as PAL2, C4H2, flavone 7-O-β-glucosyltransferase (UFGT), 4CL, C4H, chalcone synthase 2 and flavonoid 3'-monooxygenase (F3'H) genes were found to be upregulated in the senescing leaves. Biosynthesis-related transcription factors such as MYB, bHLH, and WD40 were also differentially expressed during leaf senescence[52], while bHLH, ERF, MYB, bZIP, and NAC were differentially expressed during flower growth[53]. Further analysis of the transcription factors revealed that MYB12, MYB44, MYB75, MYB114, MYC12, bHLH113, and TTG1 are crucial in luteoloside biosynthesis[52,53].
Biosynthesis of iridoids and triterpene saponins
-
The biosynthesis of terpenoids mainly involves three stages; formation of intermediates, formation of basic structural skeleton, and modification of basic skeleton[54]. The intermediates of terpenoids are mainly formed through the mevalonate (MVA) and methylerythritol phosphate (MEP) pathways, and eventually converted to the universal isoprenoid precursors, isopentenyl pyrophosphate (IPP) and its isomer dimethylallyl pyrophosphate (DMAPP) through a series of enzyme-catalyzed reactions. Under the catalysis of geranyl pyrophosphatase (GPS), IPP is then converted to geranyl pyrophosphate (GPP). Different terpenoids are subsequently derived from GPP as the intermediate product. For instance, in the formation of secoiridoid, GPP first removes the phosphoric acid group to obtain geraniol, second through a series of reactions such as oxidation and cyclization, the skeleton of iridoid, namely iridodial, can be obtained. Finally, through a series of reactions, the basic carbon skeleton of the secoiridoid, namely secologanin, is obtained[55−61] (Fig. 2c). In the formation of triterpene saponins, the key step lies in the formation of the precursor, 2,3-oxidosqualene, a reaction that is catalyzed by squalene epoxidase (SE). There are many pentacyclic triterpenes in the Lonicera genus, the most important type being the hederin-type saponins with hederagenin as aglycones. Hederin-type saponins are produced after the synthesis of oleanolic acid from β-amyrin and catalyzed by β-starch synthetase (β-AS) and Oleanolic acid synthase (OAS)[62,63]. The skeletal modification of the triterpenoid saponins is mainly achieved via the activities of the CYP450 enzymes and UDP-glycosyltransferase (UGT). Hence, the corresponding aglycones are first obtained via oxidation by the CYP450 enzymes (e.g., CYP72A), and further subjected to glycosylation by the appropriate UGT enzyme[63−65] (Fig. 2d). Skeletal formations of the iridoids and triterpene saponins in general have been well researched, but the same cannot be said about the enzymes involved in biosynthesis of these groups of compounds in the Lonicera genus. To fully utilize the iridoids and triterpene saponins in the Lonicera genus, it is necessary to further explore their biosyntheses with the view to enhancing and optimizing the process.
-
Given the importance of the bioactive compounds in the Lonicera genus, continual isolation of these compounds using the traditional methods are not only tedious and time-consuming, but also unsustainable. With the development and application of microbial metabolic engineering, different strategies have been introduced to produce these bioactive compounds by heterologous synthesis (Table 1).
Table 1. Biosynthesis of Lonicera-specialized metabolites using metabolic engineering.
Engineering bacteria Operational methods Products Yield Refs S. cerevisiae Eliminate the tyrosine-induced feedback inhibition, delete genes involved in competing pathways and overexpress rate-limiting enzymes Caffeic acid 569.0 mg/L [69] S. cerevisiae Employe a heterologous tyrosine ammonia lyase and a 4HPA3H complex composed of HpaB and HpaC derived from different species Caffeic acid 289.4 mg/L [73] S. cerevisiae Supply and recycle of three cofactors: FADH2, S-adenosyl-L-methion, NADPH Caffeic acid
Ferulic acidCaffeic acid: 5.5 g/L;
Ferulic acid: 3.8 g/L[117] E. coli Knocking out competing pathways Caffeic acid 7,922 mg/L [118] E. coli Artificial microbial community, a polyculture of three recombinant Escherichia coli strains Chlorogenic acid 250 μM [68] Cell-free biosynthesis Extract and purify spy-cyclized enzymes (CFBS-mixture) Chlorogenic acid 711.26 mg/L [70] S. cerevisiae Three metabolic engineering modules were systematically optimized: shikimate pathway and carbon distribution, branch pathways, CGA pathway genes Chlorogenic acid Flask fermentation: 234.8 mg/L;
Fed-batch fermentation:
806.8 mg/L[119] E. coli Using modular coculture engineering: construction of the defective strain improves the production and utilization of precursor substances Chlorogenic acid 131.31 mg/L [122] E. coli Introduce heterologous UDP-glucose biosynthetic genes Luteolin 34 mg/L [120] Y. lipolytica Overexpression of the key genes involved in the mevalonate pathway, the gene encoding cytochrome P450 (CYP716A12) to that encoding NADPH-P450 reductase Oleanolic acid 129.9 mg/L [85] S. cerevisiae Improve the pairing efficiency between Cytochrome P450 monooxygenase and reductase and the expression level of key genes Oleanolic acid 606.9 mg/L [121] S. cerevisiae Heterologous expression and optimization of CrAS, CrAO, and AtCPR1, and regulation of ERG1 and NADPH regeneration system Oleanolic acid 433.9 mg/L [123] Metabolic engineering of chlorogenic acid, luteolin and their glycosides
-
Due to the demand for CGA in the food, pharmaceutical, chemical, and cosmetic industries, the traditional means of obtaining the same requires a relatively longer period for plant maturation to obtain low yields of the desired product. This therefore brings into question the sustainability and efficiency of this approach. The alternative and sustainable approach has been to produce CGA using synthetic biology and metabolic engineering.
Current research has sought to utilize Escherichia coli (and its mutant strain) and Saccharomyces cerevisiae to synthetically generate CGA and other flavonoids[66−73]. For instance, Cha et al. employed two strains of E. coli to produce a relatively good yield of CGA (78 mg/L). Their approach was based on the ability of one strain to generate caffeic acid from glucose and the other strain to use the caffeic acid produced and quinic acid as starting materials to synthesize CGA[66]. Using a bioengineered mutant of E. coli (aroD mutant), Kim et al. increased the yield of CGA to as high as 450 mg/L[67]. Others have sought to increase the yield of CGA by employing a polyculture of three E. coli strains that act as specific modules for the de novo biosynthesis of caffeic acid, quinic acid and CGA. This strategy eliminates the competition posed by the precursor of CGA (i.e., caffeic acid and quinic acid) and generally results in improved production of CGA[68]. Saccharomyces cerevisiae is a chassis widely used for the production of natural substances from plants with an intimal structure that can be used for the expression of cytochrome P450 enzymes that cannot be expressed in E. coli. Researchers have used yeast to increase the production of organic acids[69]. A de novo biosynthetic pathway for the construction of CGA in yeast has been reported new cell-free biosynthetic system based on a mixture of chassis cell extracts and purified Spy cyclized enzymes were adopted by Niu et al. to a produce the highest yield of CGA reported so far up to 711.26 ± 15.63 mg/L[70].
There are many studies on the metabolic engineering for the synthesis of flavonoids, but few on luteolin and its glycosides. Strains of E. coli have been engineered with specific uridine diphosphate (UDP)-dependent glycosyltransferase (UGT) to synthesize three novel flavonoid glycosides. These glycosides were quercetin 3-O-(N-acetyl) quinovosamine (158.3 mg/L), luteolin 7-O-(N-acetyl) glucosaminuronic acid (172.5 mg/L) and quercetin 3-O-(N-acetyl)-xylosamine (160.8 mg/L)[71]. Since most of the flavonoid glycosides synthesized in E. coli are glucosylated, Kim et al. in their bid to synthesize luteolin-7-O-glucuronide, deleted the araA gene that encodes UDP-4-deoxy-4-formamido-L-arabinose formyltransferase/UDP-glucuronic acid C-4'' decarboxylase in E.coli and were able to obtain a yield of 300 mg/L of the desired product[72].
Metabolic engineering of iridoids and triterpene saponins
-
Terpenoidal saponins are mostly derived from slow-growing plants and usually possess multiple chiral centers[74]. Traditional isolation and even chemical synthesis of the terpenoidal saponins are both tedious and uneconomical for large-scale production. Therefore, it is necessary to find other ways to synthesize these compounds known to have diverse pharmacological functions.
Heterologous synthesis has become an important way to improve the target products. With the development of synthetic biology, heterologous synthesis of triterpene saponins involves chassis of both plant and microbial origin. In this regard, Nicotiana benthamiana is a model plant species for the reconstruction of the biosynthetic pathways of different bioactive compounds including monoterpenes, hemiterpenes, and diterpenes[59,75−77]. Aside from Nicotiana benthamiana, other plants have also been used as heterologous hosts[78]. Heterologous synthesis using microbial hosts mainly involves Saccharomyces cerevisiae and Escherichia coli[79−81], and other microorganisms[82,83]. Comparatively, plants as biosynthetic hosts have the advantages of an established photosynthetic system, abundant supply of relevant enzymes, and presence of cell compartments, etc. They are however not as fast growing as the microorganisms, and it is also difficult to extract and separate the desired synthesized compounds from them as hosts.
Although heterologous synthesis has many advantages, the premise of successful construction of synthetic pathway in host is to elucidate the unique structure of the compound and the key enzyme reaction mechanism in the biosynthetic pathway. There is little research on metabolic engineering of the hederin-type pentacyclic triterpene saponins in Lonicera, but there are studies on the heterologous synthesis of its aglycone precursor, oleanolic acid[84,85]. There is a dearth of scientific literature on key enzymes in the biosynthesis of pentacyclic triterpenoid saponins in the Lonicera genus.
-
Scientific evidence by diverse research groups has linked members of the Lonicera genus to a wide range of pharmacological effects (Fig. 3). These pharmacological effects are elicited by different chemical constituents, much of the underlying mechanisms of which have been elucidated by the omics techniques. Here, we summarize the pharmacological effects and pharmacodynamics of the Lonicera genus in the last 6 years.
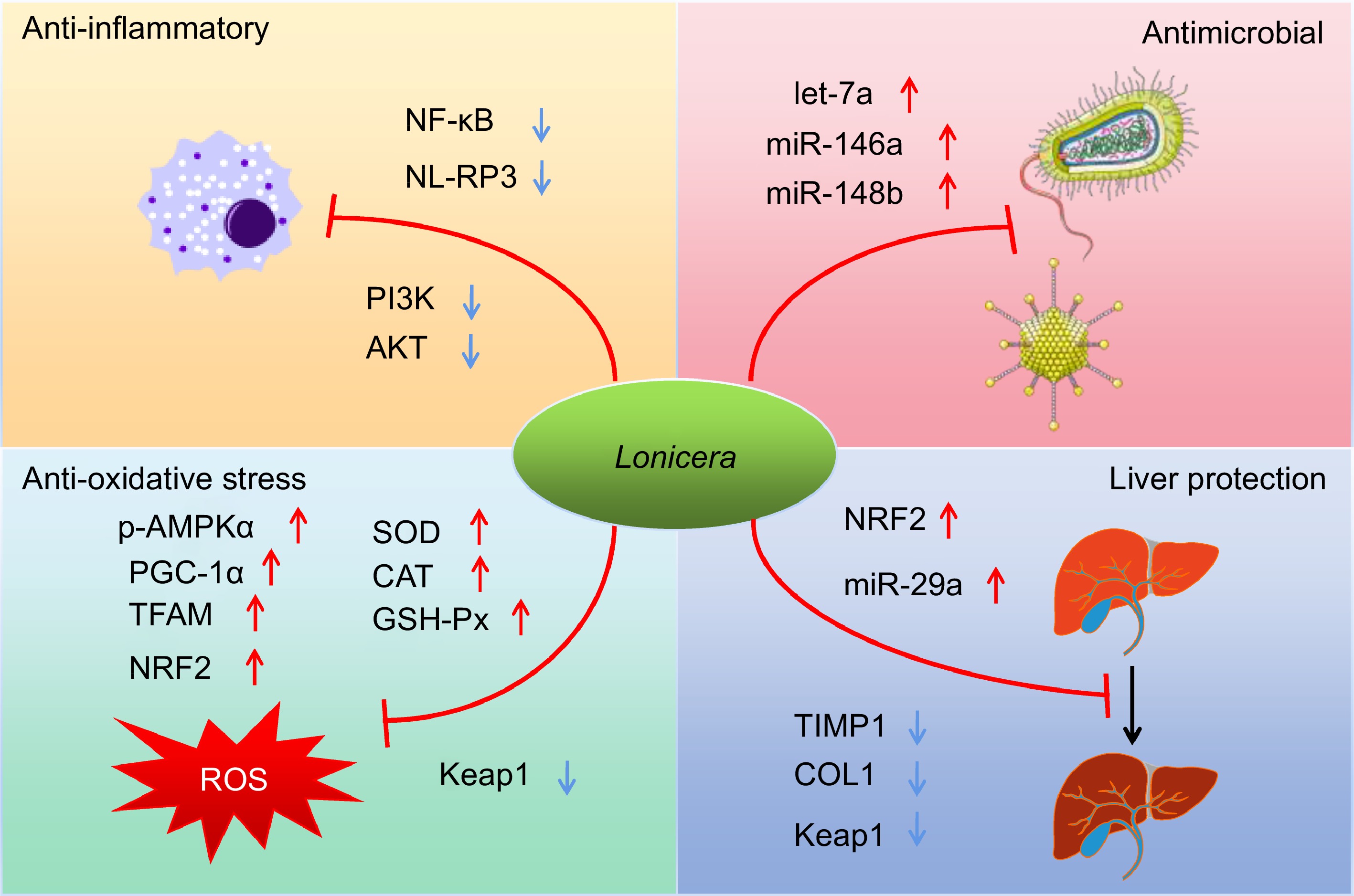
Figure 3.
Schematic summary of four main pharmacological effects (anti-inflammatory, antimicrobial, anti-oxidative and hepatoprotective effects) of the Lonicera genus and the underlying mechanisms of actions.
Anti-inflammatory effects
-
Bioactive compounds of plants in the Lonicera genus have demonstrated varying degrees of anti-inflammatory actions. In a recent study, Lv et al. showed that lonicerin inhibits the activation of NOD-like receptor thermal protein domain associated protein 3 (NLRP3) through regulating EZH2/AtG5-mediated autophagy in bone marrow-derived macrophages of C57BL/6 mice[86]. The polysaccharide extract of L. japonica reduces atopic dermatitis in mice by promoting Nrf2 activation and NLRP3 degradation through p62[87]. Several products of Lonicera have been reported to have ameliorative effects on DSS-induced colitis. Among them, flavonoids of L. rupicola can improve the ulcerative colitis of C57BL/6 mice by inhibiting PI3K/AKT, and pomace of L. japonica can improve the ulcerative colitis of C57BL/6 mice by improving the intestinal barrier and intestinal flora[88,89]. The flavonoids can also ameliorate ulcerative colitis induced by local enema of 2,4,6-trinitrobenzene sulfonic acid (TNBS) in Wistar rats by inhibiting NF-κB pathway[90]. Ethanol extract from L. Japonica has demonstrated the potential to inhibit the expressions of inflammatory cytokines in serum and macrophages of LPS-induced ICR mice[91]. The water extract of L. japonica and luteolin were found to exhibit their anti-inflammatory effects via the inhibition of the JAK/STAT1/3-dependent NF-κB pathway and induction of HO-1 expression in RAW263.7 cells induced by pseudorabies virus (PRV)[92].
Antimicrobial effects
-
Existing scientific evidence indicates that the extracts of plants in the Lonicera genus exhibit strong inhibition against different pathogenic microorganisms. Phenolic compounds from L. japonica demonstrated a particularly significant inhibitory effect against Staphylococcus aureus and Escherichia coli, in vitro, making these compounds potential food preservatives[93]. Influenza A virus is a serious threat to human health. Recent research has found the ethanol extract of L. japonica to possess a strong inhibitory effect against H1N1 influenza virus-infected MDCK cells and ICR mice[94]. The incidence of the COVID-19 pandemic called to action various scientists in a bid to find safe and efficacious treatment[95]. Traditional Chinese medicines became an attractive alternative in this search. The water extract of the flower bud of L. japonica which has traditionally served as a good antipyretic and antitussive agent attracted the attention of researchers. Scientific evidences have confirmed that the water extract of L. japonica can induce let-7a expression in human rhabdomyosarcoma cells or neuronal cells and blood of lactating mice, inhibiting the entry and replication of the virus in vitro and in vivo[96]. In addition, the water extract of L. japonica also inhibits the fusion of human lung cancer cells Calu-3 expressing ACE2 receptor and BGK-21 cells transfected with SARS-CoV-2 spike protein, and up-regulates the expression of miR-148b and miR-146a[97].
Anti-oxidative stress effect
-
Oxidative stress has been implicated in the pathophysiology of many diseases, hence, amelioration of the same could be a good therapeutic approach[98,99]. In keeping with this therapeutic strategy, various compounds from the Lonicera genus have demonstrated the ability to relieve oxidative stress due to their pronounced antioxidant effects. For instance, the polyphenolic extract of L. caerulea berry was found to activate the expression of AMPK-PGC1α-NRF1-TFAM proteins in the skeletal muscle mitochondria, improve the activity of SOD, CAT and GSH-Px enzymes in blood and skeletal muscle, relieve exercise fatigue in mice by reducing oxidative stress in skeletal muscle, and enhance mitochondrial biosynthesis and cell proliferation[100]. The diverse health benefits of the anthocyanins from L. japonica have been mainly credited to their antioxidant and anti-inflammatory effects. The anthocyanin and cyanidin-3-o-glucoside have been reported to possess the potential to prolong life and delay senescence of Drosophila through the activation of the KEAP1/NRF2 signaling pathway[101].
Hepatoprotection
-
The liver is an essential organ that contributes to food digestion and detoxification of the body. These functions expose the liver to diverse toxins and metabolites. The Lonicera genus is rich in phytochemicals that confer protection on the liver against various toxins. The phenolic compound, 4, 5-di-O-Caffeoylquinic acid methyl ester was shown to be able to improve H2O2-induced liver oxidative damage in HepG2 cells by targeting the Keap1/Nrf2 pathway[102]. Hepatic fibrosis is a complex dynamic process, with the propensity to progress to liver cancer in severe cases. The L. japonicae flos water extract solution increased the cell viability of FL83B cells treated with thioacetamide (TAA), decreased the levels of serum alanine aminotransferase (ALT) and alkaline phosphatase (ALP), inhibited the transformation growth factor β1 (TGF-β1) and liver collagen deposition[103]. Sweroside, a secoiridoid glucoside isolate of L. japonica is known to protect the C57BL/6 mice liver from hepatic fibrosis by up-regulating miR-29a and inhibiting COL1 and TIMP1[104].
Other pharmacological effects
-
Aside from the aforementioned, other pharmacological effects have been ascribed to the Lonicera genus. The ethanolic extract of L. caerulea has been reported to inhibit the proliferation of SMMC-7721 and H22 hepatoma cells, while its anthocyanins induced the apoptosis of tumor cells via the release of cytochrome C and activation of caspase[105]. AMPK/PPARα axes play an important role in lipid metabolism. A chlorogenic acid-rich extract of L. Japonica was found to significantly decrease the early onset of high-fat diet-induced diabetes in Sprague-Dawley rats via the CTRPs-AdipoRs-AMPK/PPARα axes[106]. In a high-fat diet-induced non-alcoholic fatty liver disease in C57BL/6 mice, treatment with L. caerulea polyphenol extract decreased serum inflammatory factors and endotoxin levels and the Firmicutes/Bacteroidetes ratio, an indication of its modulatory effect on the gut microbiota[107]. The iridoid-anthocyanin extract from L. caerulea berry contributed to alleviating the symptoms of intestinal infection with spirochaeta in mice[108].
-
The traditional classification of the Lonicera genus based on the morphology of member plants is further categorized into two subgenera, Chamaecerasus and Periclymenum. The Chamaecerasus includes four categories, Coeloxylosteum, Isika, Isoxylosteum and Nintooa. The Periclymenum includes two categories, Subsect. Lonicera and Subsect. Phenianthi (Supplemental Table S1).
High-throughput chloroplast genome sequencing of L. japonica found its length to be 155078 bp, which is similar to the structure of the typical angiosperm chloroplast genome. It contains a pair of inverted repeat regions (IRa and IRb, 23774 bp), a large single copy region (LSC, 88858 bp) and a small single copy area (SSC, 18672 bp)[109,110]. However, compared with chloroplast genomes of other plants, the chloroplast genome of L. japonica has a unique rearrangement between trnI-CAU and trnN-GUU[110]. Based on the phylogenetic analysis of the plastid genomes of seven plants in the Lonicera genus, 16 diverging hot spots were identified as potential molecular markers for the development of the Lonicera plants[111]. The phylogeny of Lonicera is rarely researched at the molecular level and the pattern of repetitive variation and adaptive evolution of the genome sequence is still unknown. Chloroplast genome sequences are highly conserved, but insertions and deletions, inversions, substitutions, genome rearrangements, and translocations also occur and have become powerful tools for studying plant phylogeny[112,113].
We present here the phylogenetic tree of the Lonicera genus based on the published complete chloroplast genome sequences downloaded from the National Center for Biotechnology Information (NCBI) database using the Maximum likelihood method (Fig. 4). Based on our chloroplast phylogenies, we propose to merge L. harae into Sect. Isika and L. insularis into Chamaecerasus, but whether L. insularis belongs to Sect. Isika or Sect. Coeloxylosteum is uncertain. Based on protein-coding regions (CDS) of the chloroplast genome or complete chloroplast genomes, Liu et al. and Chen et al. supported the classification of the two subgenera in Lonicera[111,114]. Sun et al. and Srivastav et al. demonstrated a classification between the two subgenera with more species by using sequences of nuclear loci generated, chloroplast genome, and restriction site-associated DNA sequencing (RADSeq)[115,116]. However, our phylogenetic analysis and that of Sun et al. show relations within the subgenus Chamaecerasus are tanglesome in some respects[116]. Plant traits are affected by the environment to varying degrees. Since evidence of plant speciation is implicit in its genome sequence, comparative analysis at the molecular level provides a relatively accurate depiction of inherent changes that might have occurred over time. These findings suggest the need for more species of the Lonicera genus to be sequenced to provide a more accurate theoretical basis for the evolution of the Lonicera plants and a more effective revision in the classification of the Lonicera genus.
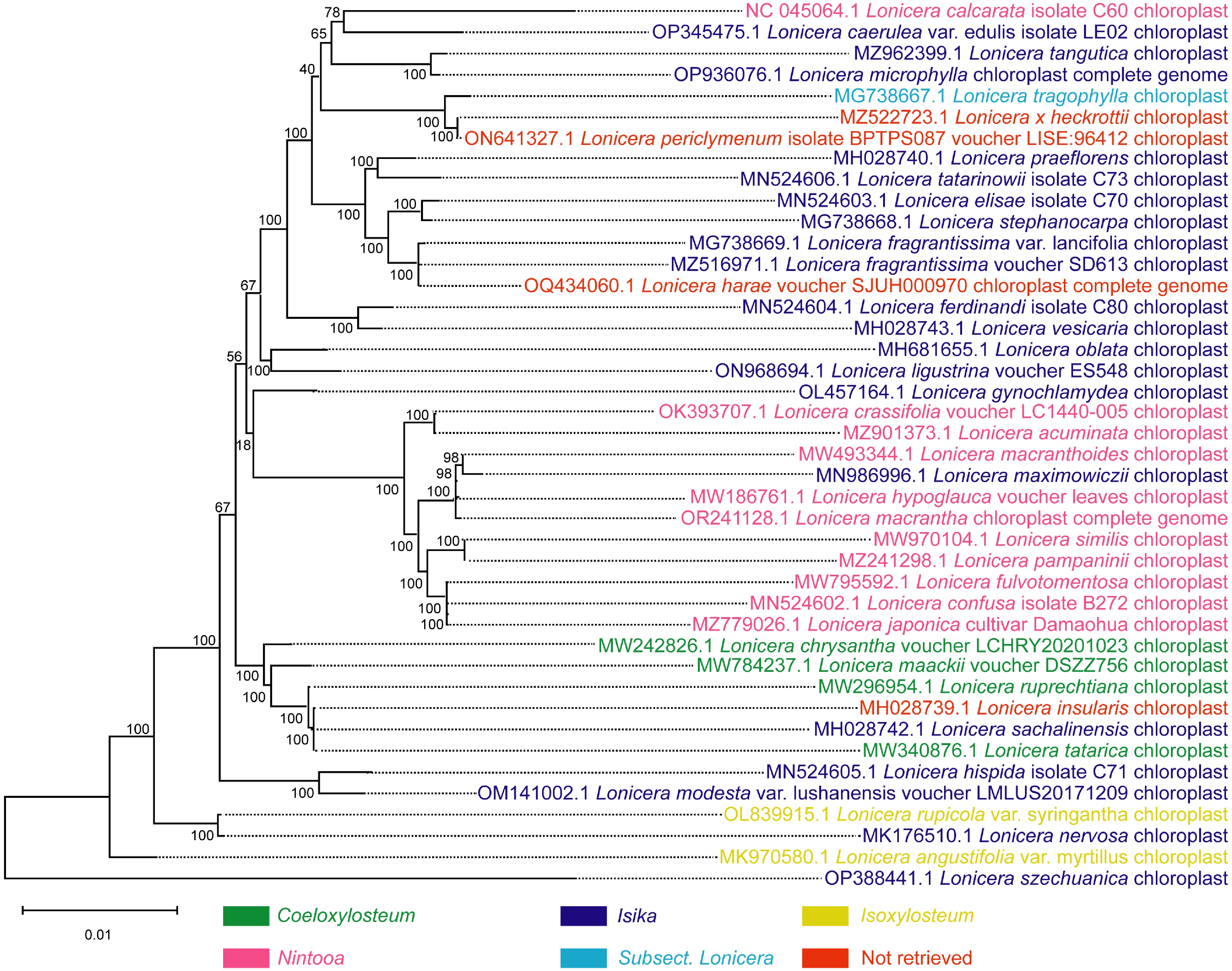
Figure 4.
Phylogenetic tree of 42 species of the Lonicera genus based on complete chloroplast genome sequence data. The phylogenetic tree was constructed by the maximum likelihood method. Coeloxylosteum, Isika, Isoxylosteum, and Nintooa belong to Chamaecerasus and Subsect. Lonicera belongs to the Periclymenum. Chamaecerasus and Periclymenum are the two subgenera of Lonicera. 'Not retrieved' indicates that the species failed to retrieve a subordinate taxon in the Lonicera.
-
The Lonicera genus is rich in diverse bioactive compounds with immeasurable prospects in many fields. Members of this genus have been used for thousands of years in traditional Chinese medicine for heat-clearing and detoxification. These plants generally have a good taste and form part of the ingredients of various fruit juices. In cosmetics, they are known to possess anti-aging and moisturizing functions. Plants of the Lonicera genus are also known for their good ecological adaptability and can be used to improve soil and ecological environment. Based on the value of the Lonicera genus, besides researching their use through molecular biological means, their efficient utilization can also be promoted in the following ways: (1) The stems and leaves of the plants could be developed for consumption and use since the chemical profiles of these parts do not differ significantly from the flowers. This way, the wastage of this scarce resource could be minimized or avoided. (2) Most of the Lonicera plants are vines or shrubs and their natural regeneration speed is slow, so the introduction and domestication of species could be strengthened to avoid overexploitation of wild resources.
At present, only the research on the biosynthesis and efficacy of chlorogenic acid is quite comprehensive and has been used widely in various fields. There is limited research on various aspects of other bioactive compounds and should therefore be given priority in future research goals. Currently, the multi-omics analytical approach has gradually evolved as a reliable and helpful analytical platform. Hence, multi-omics research on the Lonicera genus could lead to discoveries in drug discovery and human health.
-
The authors confirm contribution to the paper as follows: study conception and design, draft manuscript preparation: Yin X, Chen X, Li W, Tran LSP, Lu X; manuscript revision: Yin X, Chen X, Li W, Tran LSP, Lu X, Chen X, Yin X, Alolga RN; data/literature collection: Chen X, Yin X; figure preparation: Chen X, Yin X; figure revision: Alolga RN, Yin X, Chen X, Li W, Tran LSP, Lu X. All authors reviewed the results and approved the final version of the manuscript.
-
All data generated or analyzed during this study are included in this published article and its supplementary information file.
This work was partially supported by the National Natural Science Foundation of China (NSFC, Nos 82173918 and 82373983).
-
The authors declare that they have no conflict of interest. Xiaojian Yin is the Editorial Board member of Medicinal Plant Biology who was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of this Editorial Board member and the research groups.
- Supplemental Table S1 192 species of the Lonicera genus and GeneBank used for phylogenetic analysis.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Chen X, Li W, Lu X, Tran LSP, Alolga RN, et al. 2024. Research progress on the biosynthesis, metabolic engineering, and pharmacology of bioactive compounds from the Lonicera genus. Medicinal Plant Biology 3: e008 doi: 10.48130/mpb-0024-0008
Research progress on the biosynthesis, metabolic engineering, and pharmacology of bioactive compounds from the Lonicera genus
- Received: 20 January 2024
- Revised: 25 March 2024
- Accepted: 29 March 2024
- Published online: 16 May 2024
Abstract: The Lonicera genus with medicinal and edible characteristics is a rich source of bioactive compounds that together account for numerous health benefits. Plants within this genus have been used traditionally in China to treat heat-related illnesses, viral respiratory infections, skin diseases, and inflammation for thousands of years. The main bioactive compounds include chlorogenic acids, flavonoids, iridoids, and triterpene saponins. Aside from the role of this genus as a valuable medicinal resource, constituent members have also been used in the food and cosmetic industries. Due to the huge demand for plants within the Lonicera genus, and the propensity for overexploitation of the wild resources of Lonicera, there is a need to meet this demand via alternative and efficient strategies such as metabolic engineering and green production. Here, we provide a comprehensive review of relevant scientific literature covering the structure, biosynthesis, metabolic engineering, pharmacology and phylogenetic analysis of the main bioactive constituents of the Lonicera genus. Finally, we proffer suggestions on the prospects of fully exploiting and utilizing plants of the Lonicera genus as useful medicinal plant resources.
-
Key words:
- Lonicera /
- Biosynthesis /
- Metabolic engineering /
- Pharmacology













