-
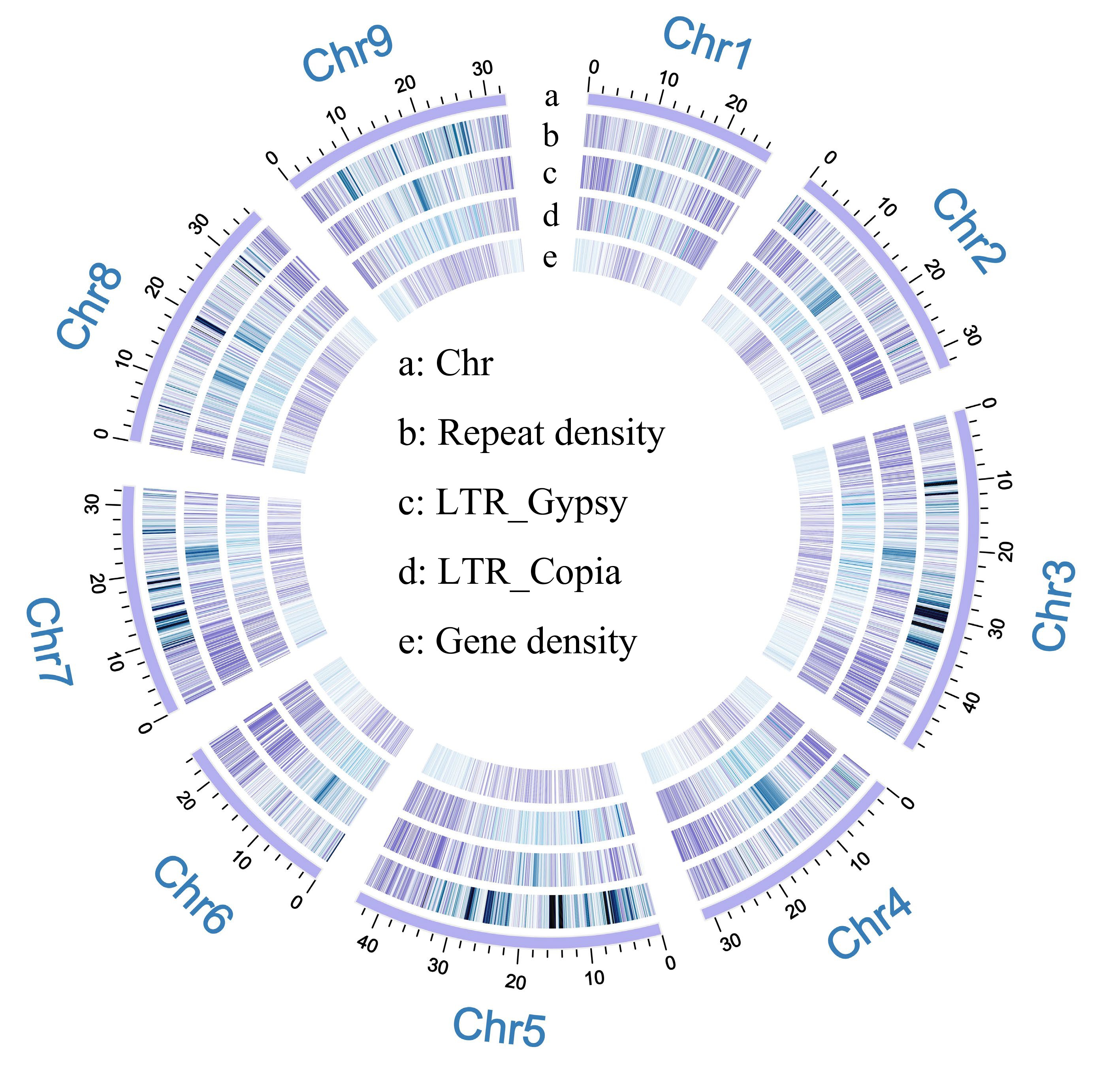
Figure 1.
Genome mapping of finger lemon. (a) Chromosomes. (b) Repeat density. (c) LTR_Gypsy (LTRs). (d) LTR_Copia (LTRs). (e) Gene density.
-
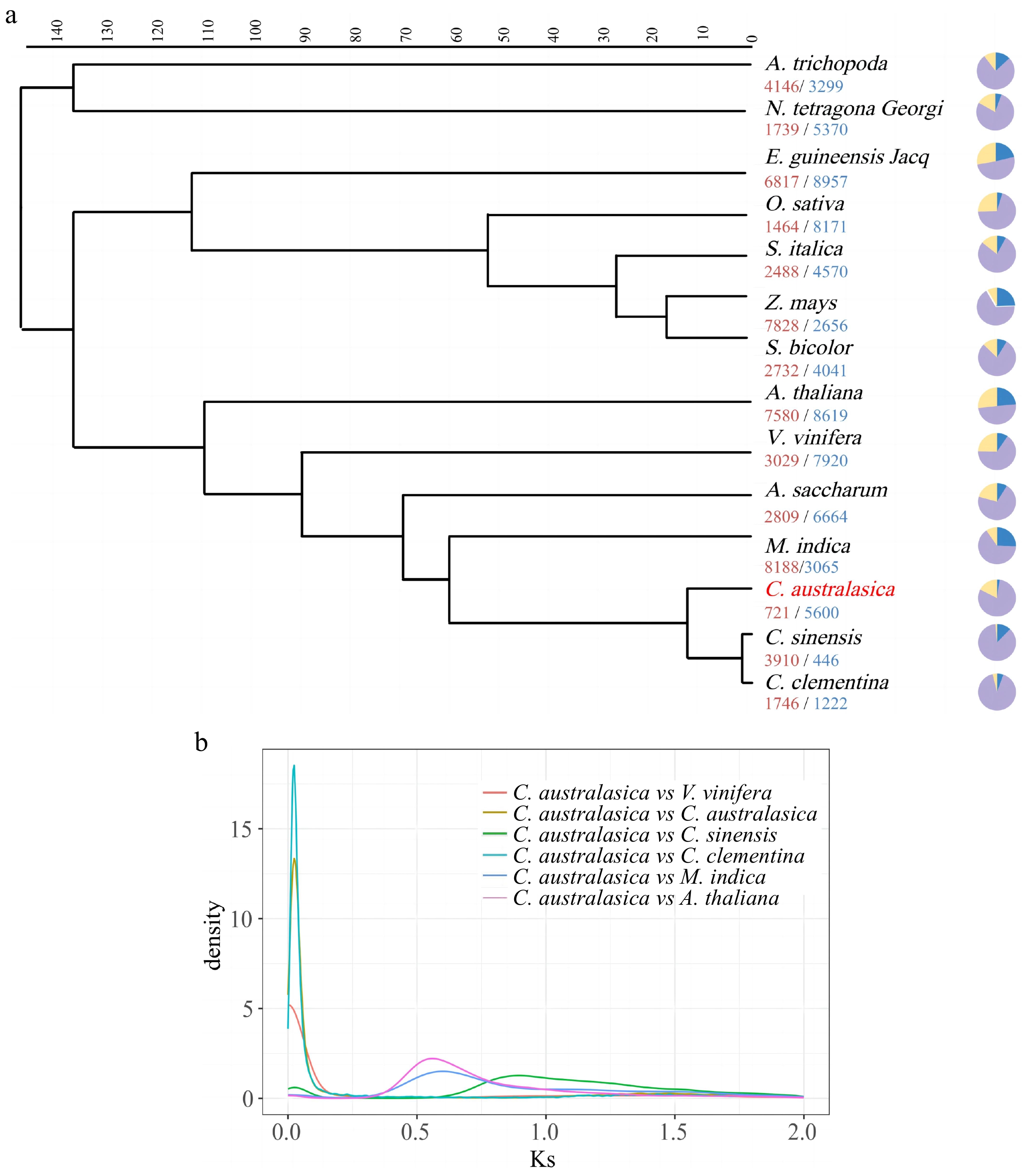
Figure 2.
Phylogeny and evolution of Citrus australasica. (a) Phylogenetic analysis based on homologous genes from 14 plant species, including five Saprotaceae species and two outgroups (Amborella trichopoda and Nymphaea L.), using the Orthfinder module. Pie charts represent the number of gene family expansions and contractions. (b) Frequency distribution of synonymous substitution rates (Ks) between homologous gene pairs in the syntenic blocks of C. australasica vs V. vinifera, C. australasica vs C. australasica, C. australasica vs C. sinensis, C. australasica vs C. clementina, C. australasica vs M. indica, C. australasica vs A. thaliana.
-
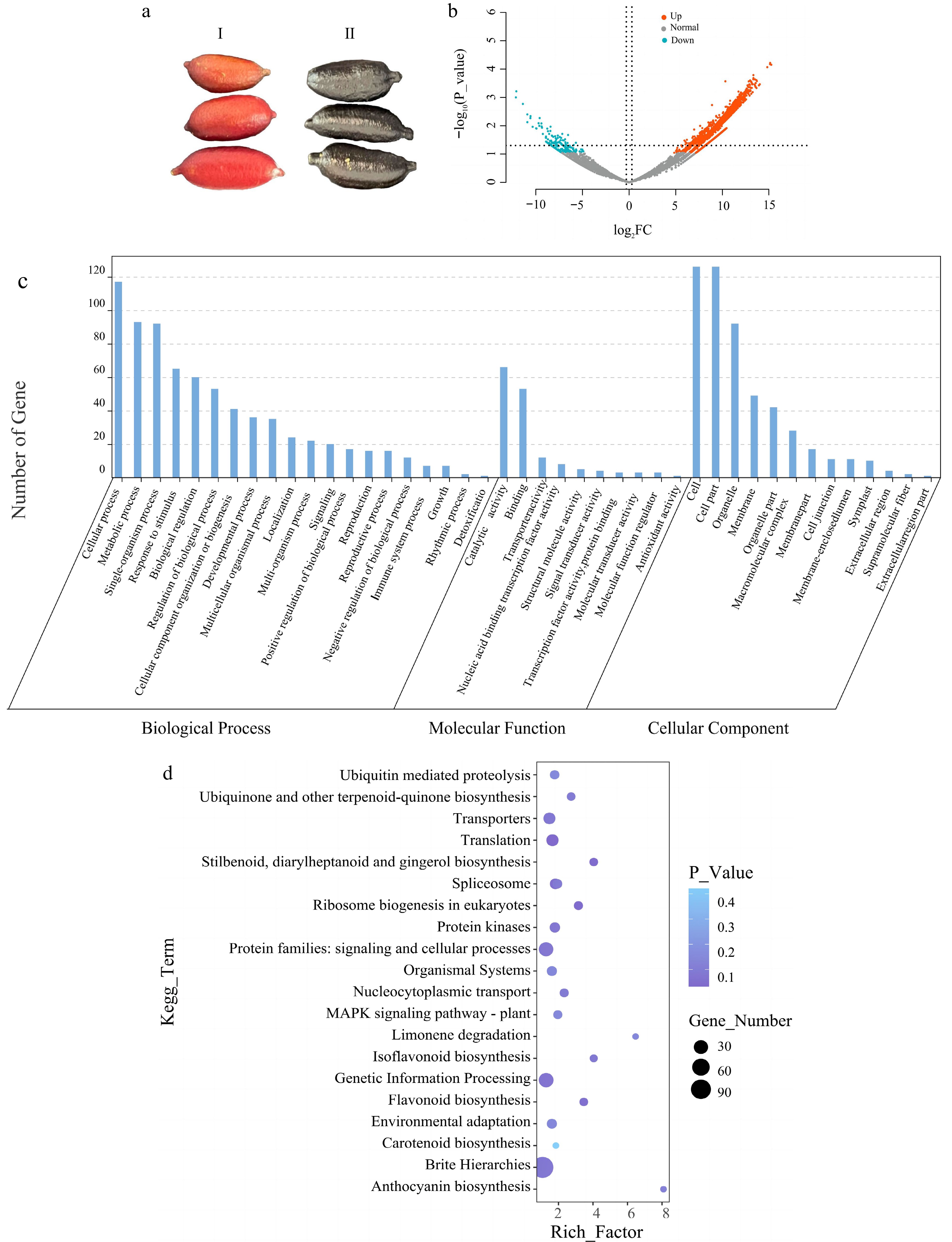
Figure 3.
Differential gene analysis of finger lemons. (a) Photographs of two finger lemon samples of different varieties 'JiaLiHong' and 'YaoJi' used for transcriptome analysis; I: 'JiaLiHong', II: 'YaoJi'. (b) Volcano plot of the expression of two different varieties of finger lemons after differential gene analysis; red indicates up-regulation; blue indicates down-regulation. (c) Enrichment results of up-regulated genes in the GO database. (d) Enrichment results of up-regulated genes in the KEGG database.
-
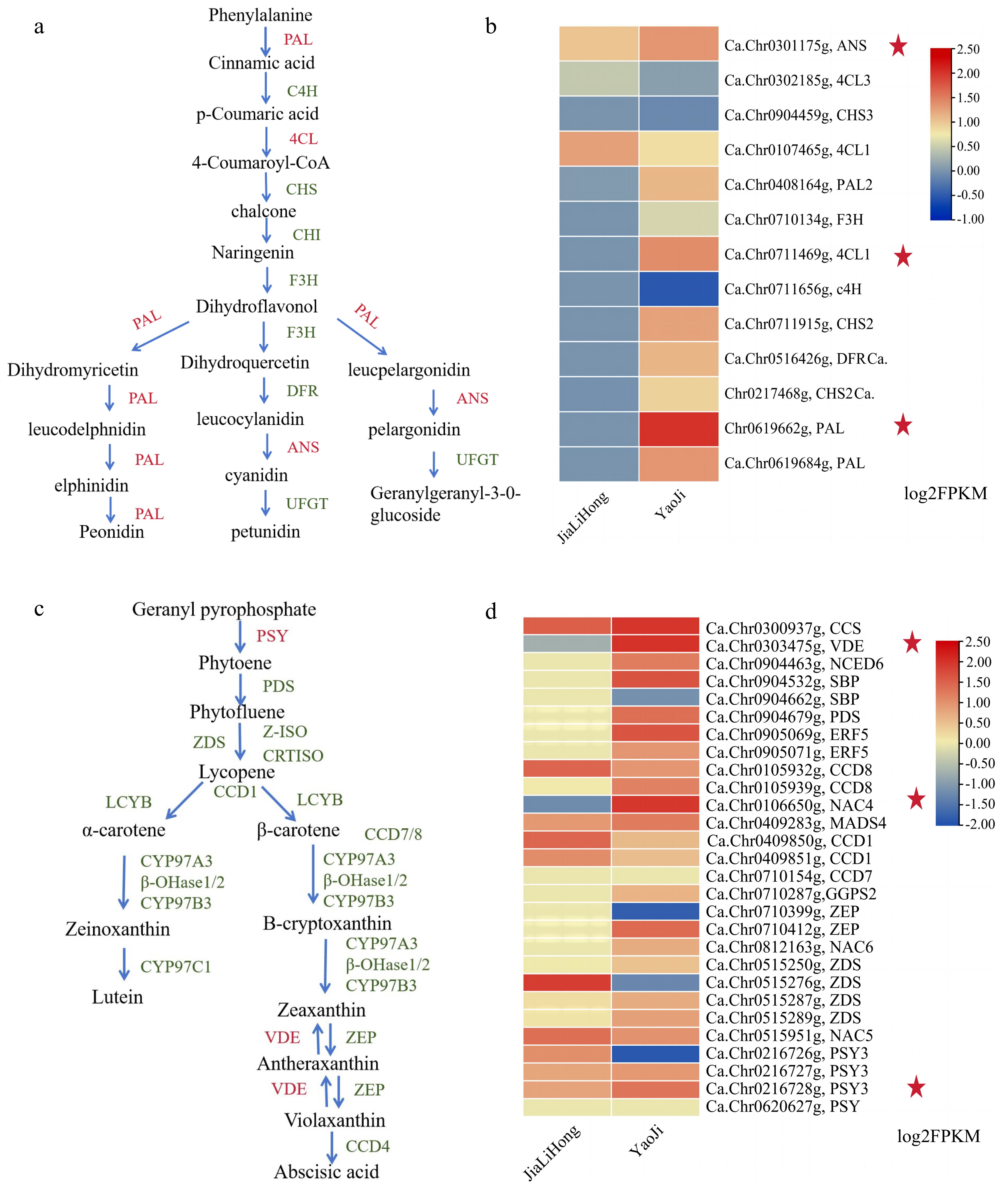
Figure 4.
Biosynthetic pathways of flavonoids and carotenoids. Normalized FPKM values were expressed using 'JiaLiHong' as control. (a) Flavonoid biosynthetic pathway. (b) Heat map of highly expressed genes for flavonoid biosynthesis in 'YaoJi'. (c) Carotenoid biosynthetic pathway d. Heat map of highly expressed genes for carotenoid biosynthesis in 'YaoJi'.
-
Genomic feature Value Genome size (Mb) 313,916,163 Chromosome-scale scaffolds 314,637,567 Total number of chromosomes 9 Number of contigs 175 Number of contigs N50 15 N50 of contigs (bp) 6,642,737 Number of contigs N80 37 Number of contigs N90 57 Total number of scaffolds 28 N50 of scaffold (bp) 32,805,073 N60 of scaffold (bp) 32,750,318 N70 of scaffold (bp) 32,749,230 N80 of scaffold (bp) 31,439,114 N90 of scaffold (bp) 27,021,073 N100 of scaffold (bp) 20,247 SD length of scaffold (bp) 313,375,644 GC content of the genome (%) 34.26% Complete BUSCOs 98.4% Quality value (QV) 27.68 LTR Assembly Index (LAI) 15.19 Repeat sequences (Gb) 164,723,365 (52.35%) Total number of protein-coding genes 21,154 Table 1.
Features of Citrus australasica genome assembly.
Figures
(4)
Tables
(1)