-
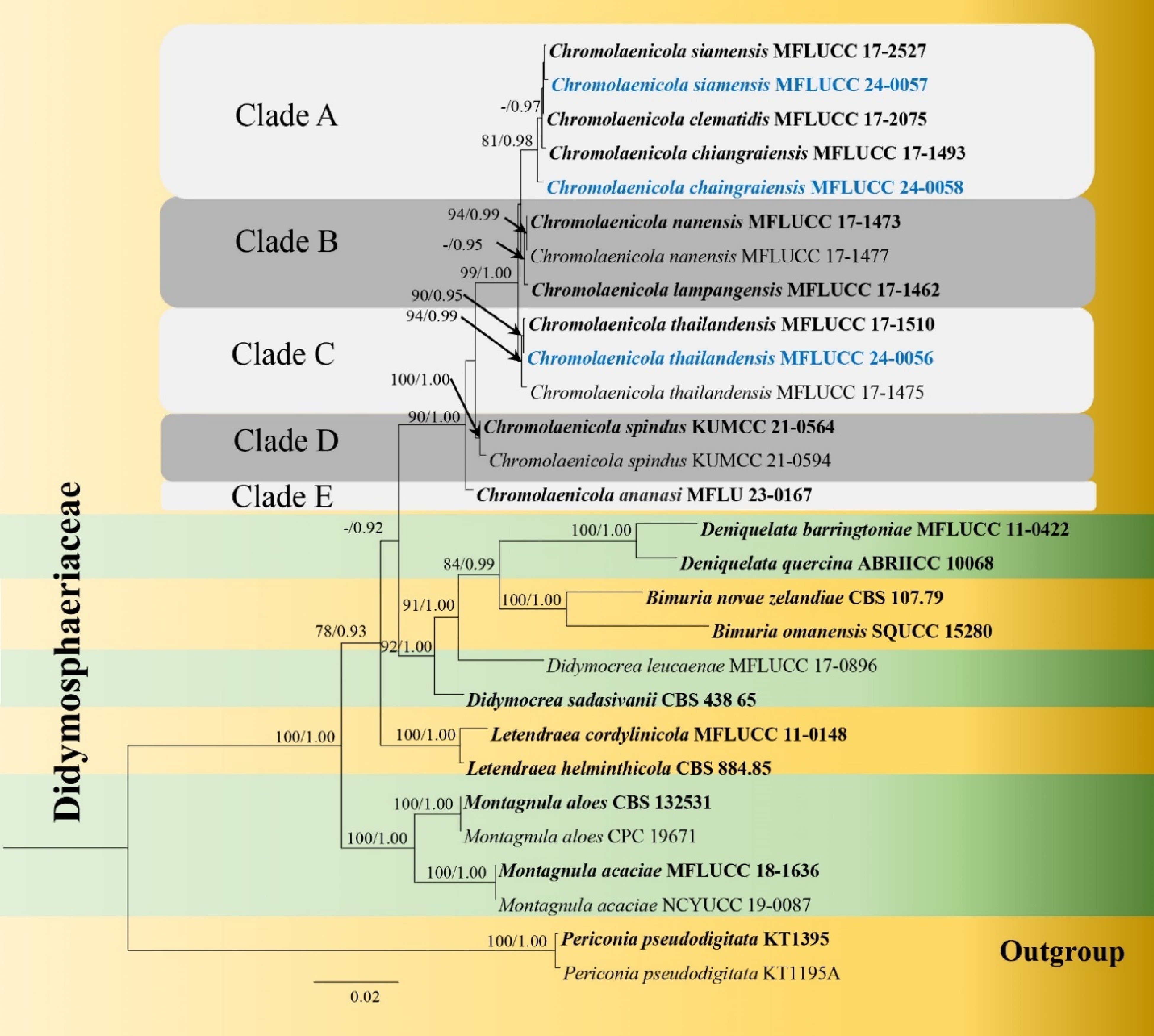
Figure 1.
Phylogram generated from maximum likelihood analysis based on the combined dataset of LSU, SSU, ITS, tef1-α and rpb2 sequence data. Bootstrap support values for ML equal to or greater than 75% and BYPP equal to or greater than 0.95 are given at the nodes. Newly generated sequences are in blue and type species are in bold.
-
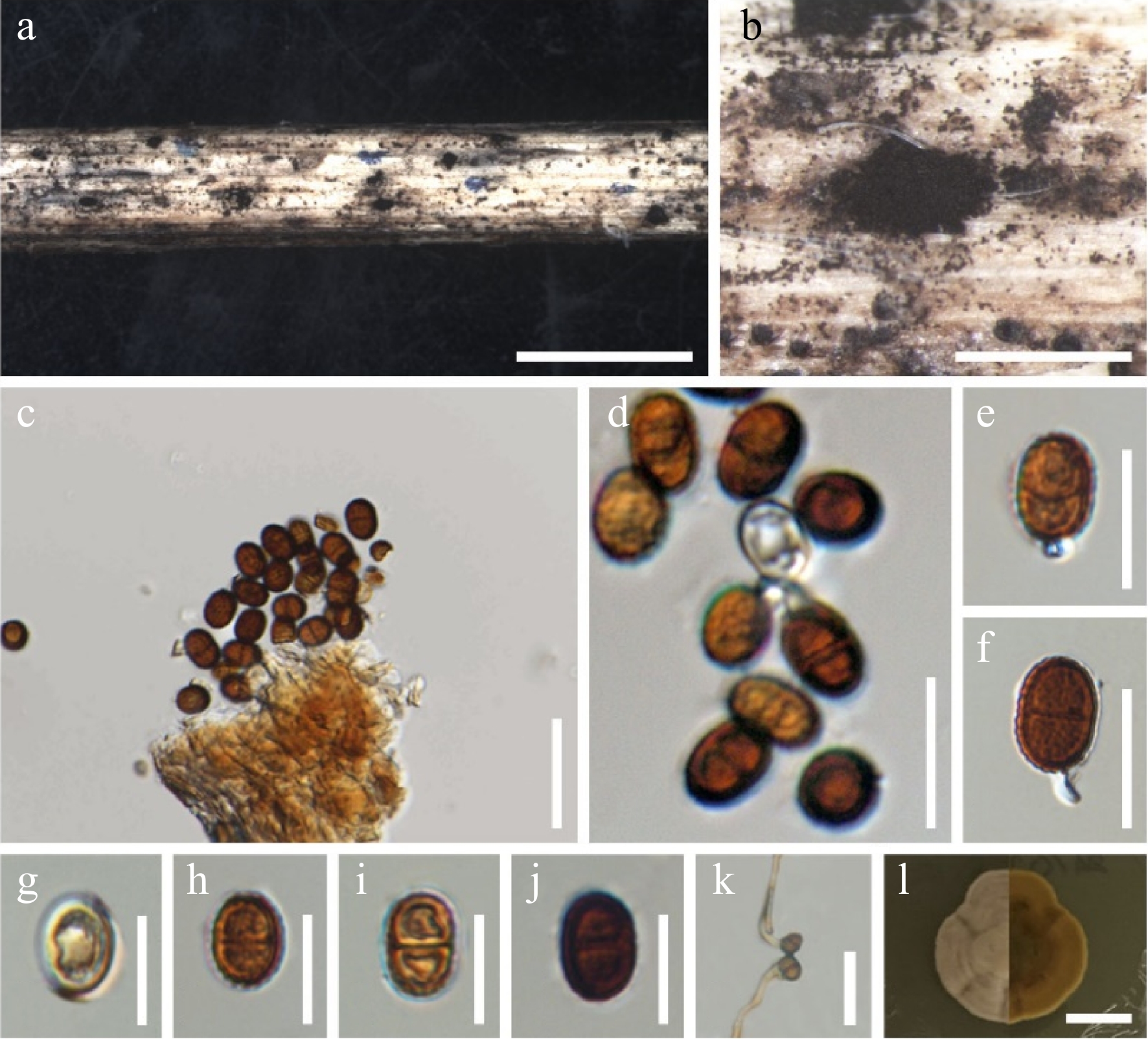
Figure 2.
Chromolaenicola chaingraiensis (MFLU 24-0030, new host record). (a), (b) Colonies on the substrate. (c)–(f) Conidia and conidiogenous cells. (g)–(j) Conidia. (k) Germinating conidia. (l) Culture on MEA. Scale bars: (a), (b) = 500 µm, (c) = 30 µm, (d)–(j) = 10 µm, (k) = 20 µm, (l) = 10 mm.
-
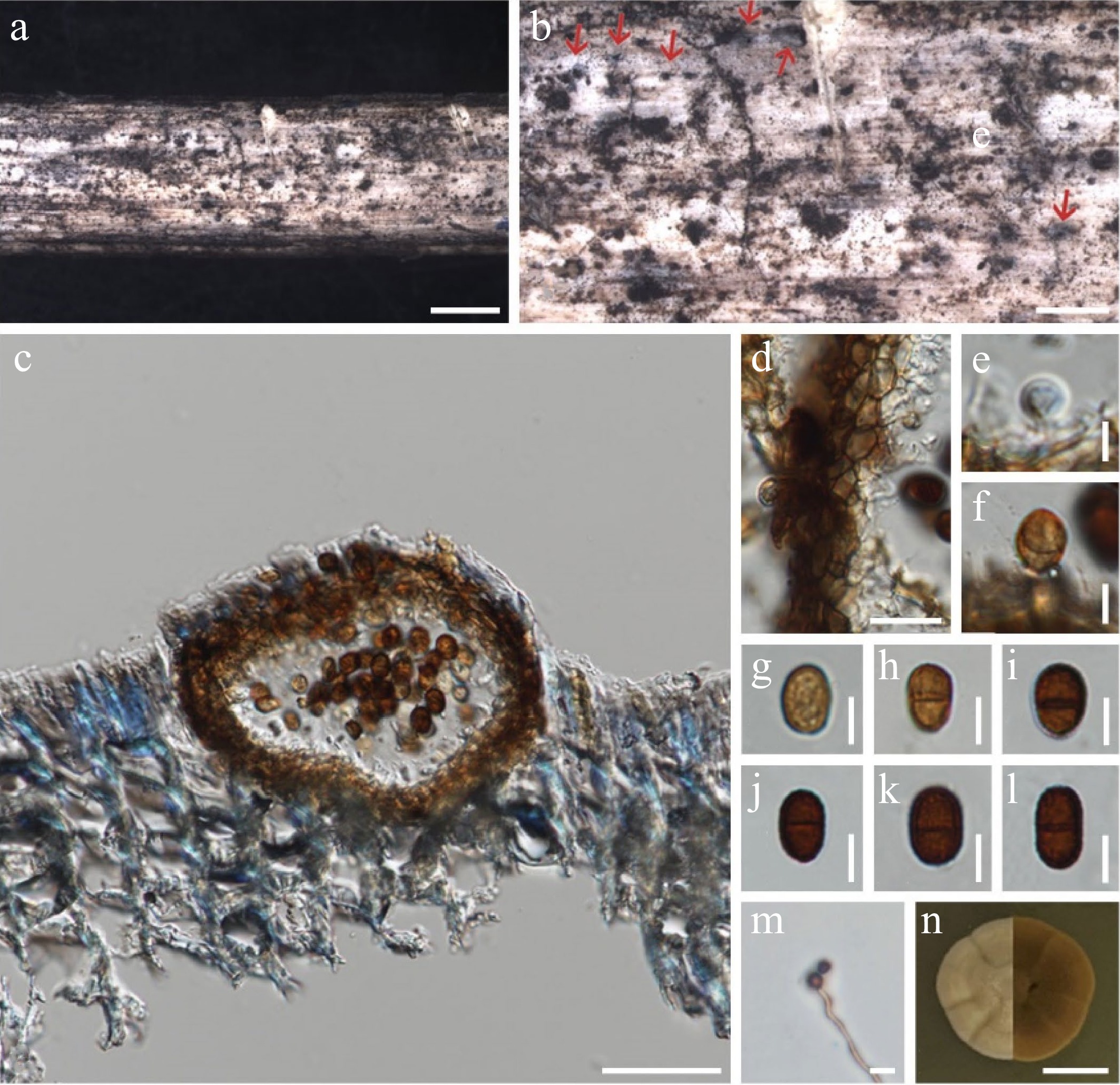
Figure 3.
Chromolaenicola siamensis (MFLU 24-0029, new host record). (a), (b) Conidiomata on the substrate. (c) Section through conidiomata. (d) Peridium. (e), (f) Conidiogenous cells. (g)–(l) Conidia. (m) Germinating conidia. (n) Culture on MEA. Scale bars: (a), (b) = 500 µm, (c) = 50 µm, (d) = 20 µm, (e)–(m) = 5 µm, (n) = 10 mm.
-
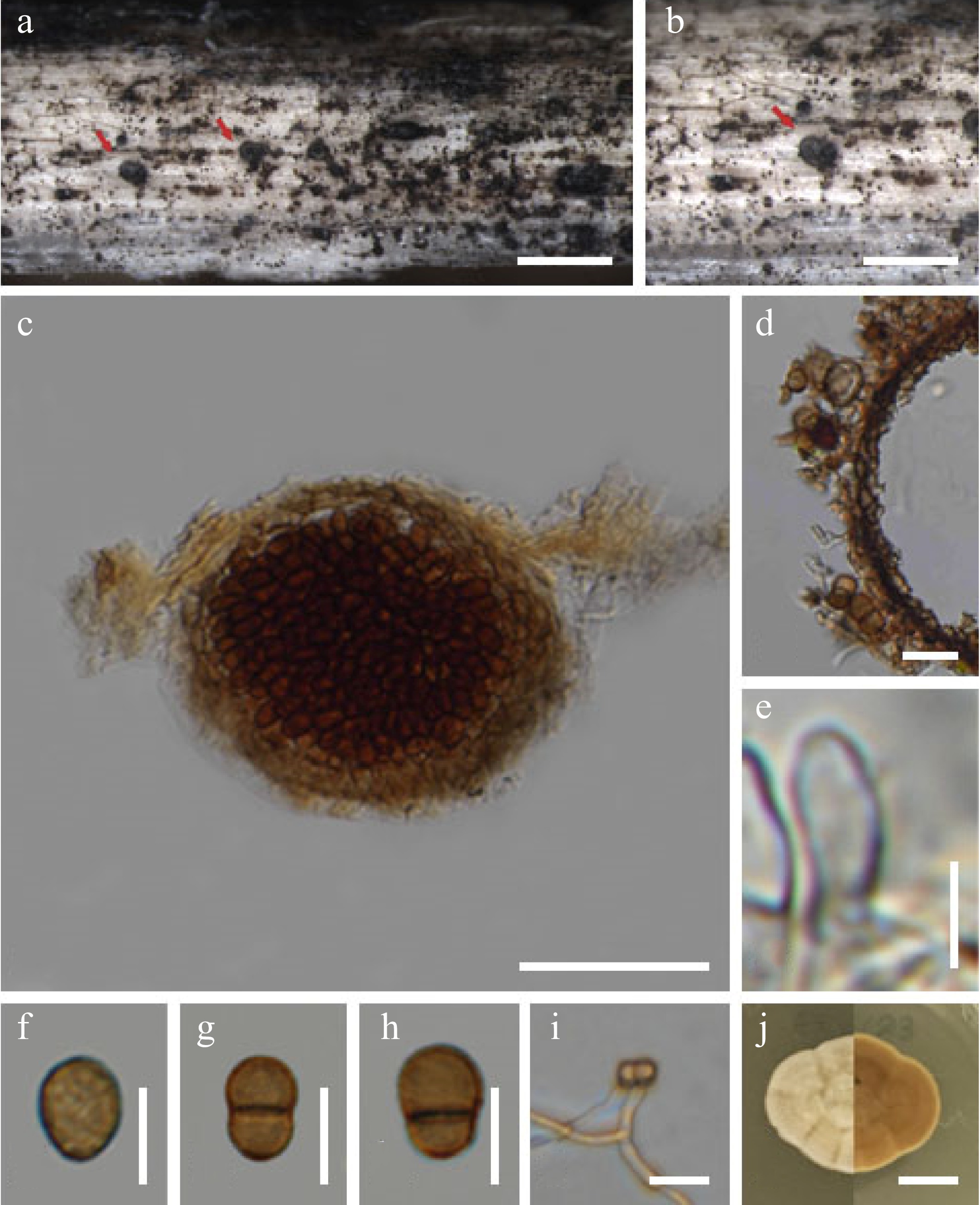
Figure 4.
Chromolaenicola thailandensis (MFLU 24-0028, new host record, first report of asexual morph). (a), (b) Colonies on substrate. (c) Section through conidiomata. (d) Peridium. (e) Conidiogenous cells. (f)–(h) Conidia. (i) Germinating conidia. (j) Culture on MEA. Scale bars: (a), (b) = 500 µm, (c) = 50 µm, (d)–(i) = 10 µm, (j) = 10 mm.
-
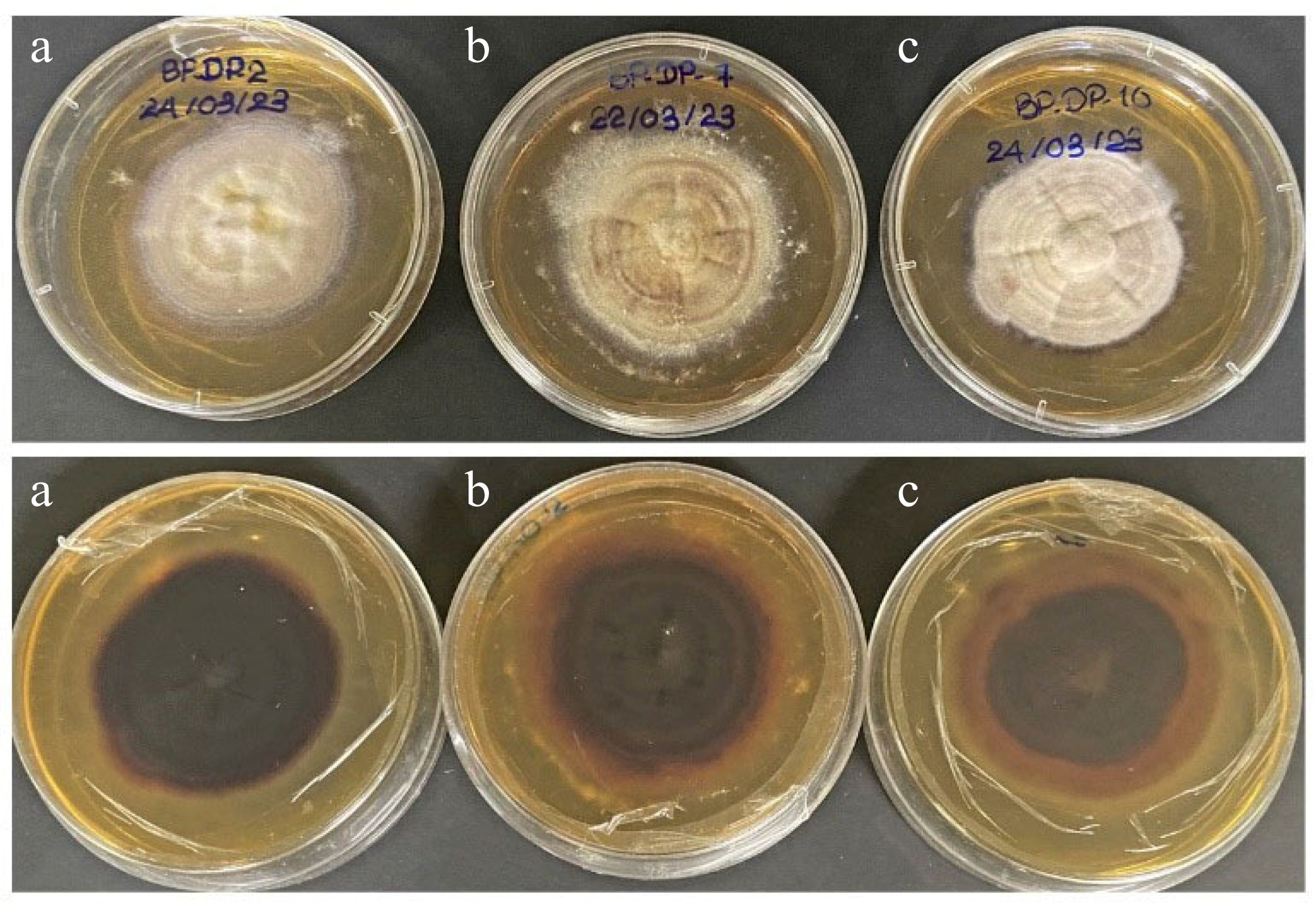
Figure 5.
Culture characteristics on MEA. (a) Chromolaenicola thailandensis (MFLUCC 24-0056), (b) Chromolaenicola siamensis (MFLUCC 24-0057), (c) Chromolaenicola chiangraiensis (MFLUCC 24-0058).
-
Gene Primers PCR conditions Ref. Forward Reverse Large subunit (LSU) LR0R LR5 95 °C: 3 min, (94 °C: 30 s, 56 °C: 50 s, 72 °C: 1 min) × 40 cycles 72 °C: 7 min [35] Small subunit (SSU) NS1 NS4 95 °C for 3 min, (94 °C: 30 s, 55 °C: 50 s, 72 °C :1 min) × 40 cycles 72 °C: 7 min [36] Internal transcribed spacer (ITS) ITS5 ITS4 95 °C for 3 min, (94 °C: 30 s, 55 °C: 50 s, 72 °C :1 min) × 40 cycles 72 °C: 7 min [36] Elongation factor-1 alpha (tef1- α) EF-1 983F EF1-2218R 95 °C: 3 min, (94 °C: 30 s, 55 °C: 50 s, 72 °C: 1 min) × 40 cycles 72 °C: 7 min [37] RNA polymerase II subunit (rpb2) fRPB2-5 F fRPB2-7cR 95 °C: 5 min, (95 °C :1 min, 52 °C: 2 min, 72 °C: 90 s) × 40 cycles 72 °C: 10 min [38] Table 1.
PCR conditions used in this study.
-
Species Strain numbers GenBank accession numbers Ref. LSU SSU ITS tef1-α rpb2 Bimuria omanensis SQUCC 15280 NG_071257 N/A NR_173301 MT279046 N/A [39] B. novae-zelandiae CBS 107.79 MH872950 NA MH861181 NA N/A [40] Chromolaenicola ananasi MFLU 23-0167 OR438811 OR458332 OR438340 OR500305 N/A [30] C. clematidis MFLUCC 17-2075T MT310601 MT214554 MT226671 N/A N/A [29] C. chiangraiensis MFLUCC 17-1493 MN325005 MN325011 MN325017 MN335650 MN335655 [17] C. chiangraiensis MFLUCC 24-0058 PP464125 PP464129 PP464138 PP474193 PP474190 This study Chromolaenicola nanensis MFLUCC 17-1477 MN325002 MN325008 MN325014 MN335647 MN335653 [17] C. nanensis MFLUCC 17-1473 MN325003 MN325009 MN325015 MN335648 MN335653 [17] C. lampangensis MFLUCC 17-1462 MN325004 MN325010 MN325016 MN335649 MN335654 [17] C. siamensis MFLUCC 17-2527 NG_066311 N/A NR_163337 N/A N/A [28] C. siamensis MFLUCC 24-0057 PP464124 PP464128 PP464137 PP474192 PP474189 This study C. sapindi KUMCC 21-0564T OP059009 OP059058 OP058967 OP135943 N/A [27] C. sapindi KUMCC 21-0594 OP059010 OP059059 OP058968 OP135944 N/A [27] C. thailandensis MFLUCC 17-1510 MN325006 MN325012 MN325018 MN335651 N/A [17] C. thailandensis MFLUCC 17-1475 MN325007 MN325013 MN325019 MN335652 MN335656 [17] C. thailandensis MFLUCC 24-0056 PP464123 PP464127 PP464136 PP474191 PP474188 This study Deniquelata barringtoniae MFLUCC 11−0422 JX254655 JX254656 NR_111779 N/A N/A [41] D. quercina ABRIICC 10068 MH316157 MH316155 MH316153 N/A N/A [42] Didymocrea leucaenae MFLUCC 17−0896 NG_066304 MK347826 NR_164298 MK360052 N/A [28] D. sadasivanii CBS 438.65 DQ384103 DQ384066 MH870299 N/A N/A [40] Letendraea cordylinicola MFLUCC 11−0148 NG_059530 NG_068362 NR_154118 N/A N/A [41] L. helminthicola CBS 884.85 AY016362 AY016345 MK404145 MK404174 N/A [43] Montagnula acaciae MFLUCC 18−1636 ON117298 ON117267 ON117280 ON158093 N/A [44] M. acaciae NCYUCC 19−0087 ON117299 ON117268 ON117281 ON158094 N/A [44] M. aloes CPC 19671 JX069847 N/A JX069863 N/A N/A [45] M. aloes CBS 132531 NG_042676 N/A NR_111757 N/A N/A [40] Periconia pseudodigitata KT1395 AB807564 AB797274 LC014591 AB808540 N/A [46] P. pseudodigitata KT1195A AB807563 AB797273 LC014590 AB808539 N/A [46] T: Type strains; Abbreviations of culture collections: CBS: Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands, CPC: Working collection of Pedro Crous housed at CBS, KT: K. Tanaka, MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand, NCYUCC: National Chiayi University Culture Collection, Taiwan. SQUCC: Sultan Qaboos University Culture Collection, Sultanate of Oman. ABRIICC: Agricultural Biotechnology Research Institute of Iran Culture Collection, Iran. Sequences generated in the current study are in bold. N/A: Not available. Table 2.
List of taxa, specimens and sequences used in phylogenetic analyses.
-
Species Conidiomata
(µm)Peridium
(µm)Conidiogenous cells
(µm)Conidia
(µm)Host/substrate Ref. C. ananasi (MFLU 23-0167) − − − 7–8 × 4–5 Ananas comosus (Bromeliaceae) [30] C. chiangraiensis
(MFLUCC 17-1493)− − 3.5–6.5 × 1–2 9–14 × 6–9 Chromolaena odorata (Asteraceae) [17] C. chaingraiensis
(MFLUCC 24-0058)− − 2–4 × 1–2 9–13 × 6–10 Bidens pilosa (Asteraceae) This study C. clematidis
(MFLUCC 17–2075)76–145 × 107–128 5–10 2.6–4.5 × 4–7 7–10 × 4.5–7 Clematis subumbellata (Ranunculaceae) [29] C. lampangensis
(MFLUCC 17-1462)150–230 × 170–270 10–20 – 12–15 × 4–6.5 Chromolaena odorata (Asteraceae) [17] C. siamensis
(= Cylindroaseptospora siamensis, MFLUCC 17–2527)110–165 × 140–190 15–38 6.5–7.4 × 3.2–4.7 7.2–9.4 × 5.4–6.5 Leucaena sp. (Fabaceae) [28] C. siamensis
(MFLUCC 24-0057)130–235 × 170–230 15–20 1–2.5 × 2–4 7–15 × 5–10 Bidens pilosa (Asteraceae) This study C. thailandensis
(MFLUCC 24-0056)100–150 × 110–150 13–20 1–2 × 3–4 5–11 × 4–10 Bidens pilosa (Asteraceae) This study Table 3.
Synopsis of recorded asexual morph of Chromolaenicola species.
-
Species Ascomata
(μm)Peridium
(μm)Asci
(μm)Ascospores
(μm)Host/substrate Ref. C. nanensis (MFLUCC 17-1473) 210–230 × 200–220 15–20 110–145 × 10–12.5 16–20 × 7.5–9 Chromolaena odorata (Asteraceae) [17] C. spindi (KUMCC 21-0564) 420–530 × 270–350 15–25 125–155 × 12–16 16–23 × 6.5–9.5 Sapindus rarak (Spindaceae) [27] C. thailandensis (MFLUCC 17-1510) 145–225 × 175–240 10–20 90–160 × 10–14 16–24 × 9–11 Chromolaena odorata (Asteraceae) [17] Table 4.
Synopsis of recorded sexual morph of Chromolaenicola species.
-
Species Zone of inhibition (mm); Ampicillin (+) Ref. Bacillus subtilis Escherichia coli Staphylococcus aureus Mucor plumbeus Chromolaenicola ananasi (MFLU 23-0167) N/A N/A N/A N/A [30] C. chaingraiensis (MFLUCC 24-0058) 10 mm inhibition no inhibition No inhibition N/A This study C. chiangraiensis (MFLUCC 17-1493) No inhibition No inhibition N/A no inhibition [17] C. clematidis (MFLUCC 17-2075) N/A N/A N/A N/A [29] C. lampangensis (MFLUCC 17-1462) No inhibition no inhibition N/A 14 mm inhibition [17] C. nanensis (MFLUCC 17-1473) No inhibition No inhibition N/A 12 mm inhibition [17] C. siamensis (MFLUCC 17-2527) N/A N/A N/A N/A [28] C. siamensis (MFLUCC 24-0057) 18 mm inhibition no inhibition no inhibition N/A This study C. spindii (KUMCC 21-0564) N/A N/A N/A N/A [27] C. thailandensis (MFLUCC 17-1510) No inhibition No inhibition N/A No inhibition [17] C. thailandensis (MFLUCC 24-0056) 17 mm inhibition No inhibition No inhibition N/A This study N/A: Not available; Positive control (+): Ampicillin. Table 5.
Preliminary antimicrobial activity result of Chromolaenicola species.
Figures
(5)
Tables
(5)