-
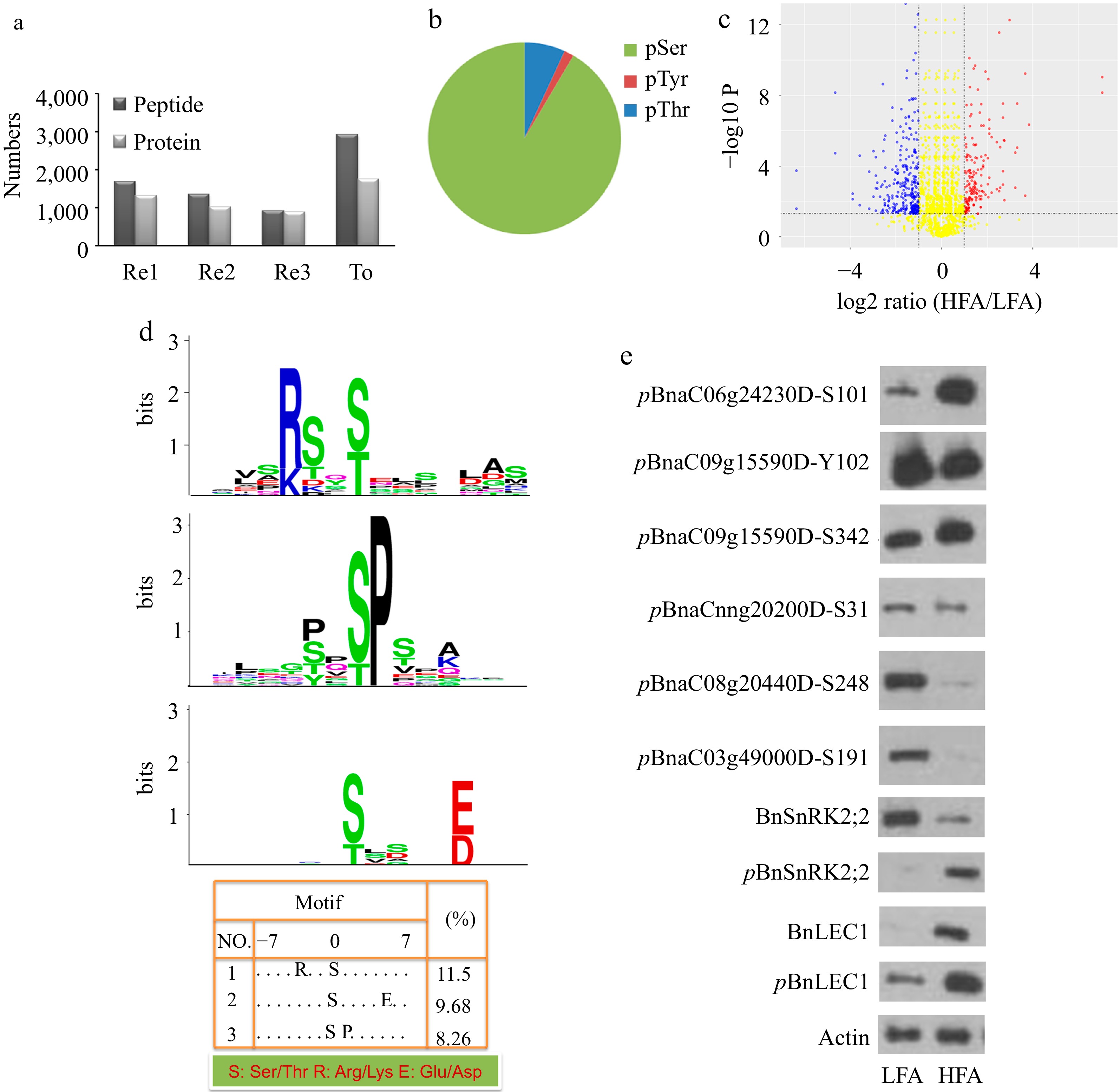
Figure 1.
Quantitative phosphoproteomic identification of phosphoproteins related oil biosynthesis and accumulation in seeds of B. napus. (a) The number of unique phosphopeptides and phosphoproteins identified from each experimental replicate (Re1, Re2, Re3) and combination of all replicates (To). Each experimental replicate comprises the mixture of dimethyl- and dimethyl:2H(6)-labeled peptides. (b) Distribution of phosphorylation sites by amino acids. (c) Volcano plots of quantitative phosphopeptide analysis. The red and blue circles represent significantly increased and decreased phosphopeptides (HFA vs. LFA), respectively. The log2 ratio represents the average binary logarithmic ratio of MS1 dimethyl-labeled areas of a phosphopeptide between HFA and LFA, P is the P value determined using Student's t test followed by Benjamini–Hochberg multiple hypothesis test correction. The horizontal and vertical dashed line indicates the threshold for the significance (FDRB.–H. ≤ 5%) and fold change (log2 ratio ≤ −1 or > 1), respectively. (d) Phosphorylation motif analysis of upregulated phosphopeptides. Phosphorylation motifs were searched by Motif-X and depicted using LOGO by MEME. The numbers and detailed sequences of phosphopeptides in each LOGO are shown in bottom right table and Supplemental Table S1− S3, respectively. (e) Immunoblotting analysis for validating the quantitative phosphoproteomic results. Protein immunoblotting analysis of protein phosphorylation in 25 DAP seeds of HFA and LFA was performed using polyclonal antibodies against anti-pBnSnRK2;2, anti-BnSnRK2;2, anti-pBnLEC1, anti-BnLEC1, anti-pBnaC06g24230DS101, anti-pBnaC09g15590DY102, anti-pBnaC09g15590DS342, anti-pBnaCnng20200DS31, anti-pBnaC08g20440DS248, and anti-pBnaC03g49000DS191. Anti-actin was used as the internal reference. HFA, a B. napus variety with high seed oil content; LFA, a B. napus variety with low seed oil content; DAP, day after pollination.
-
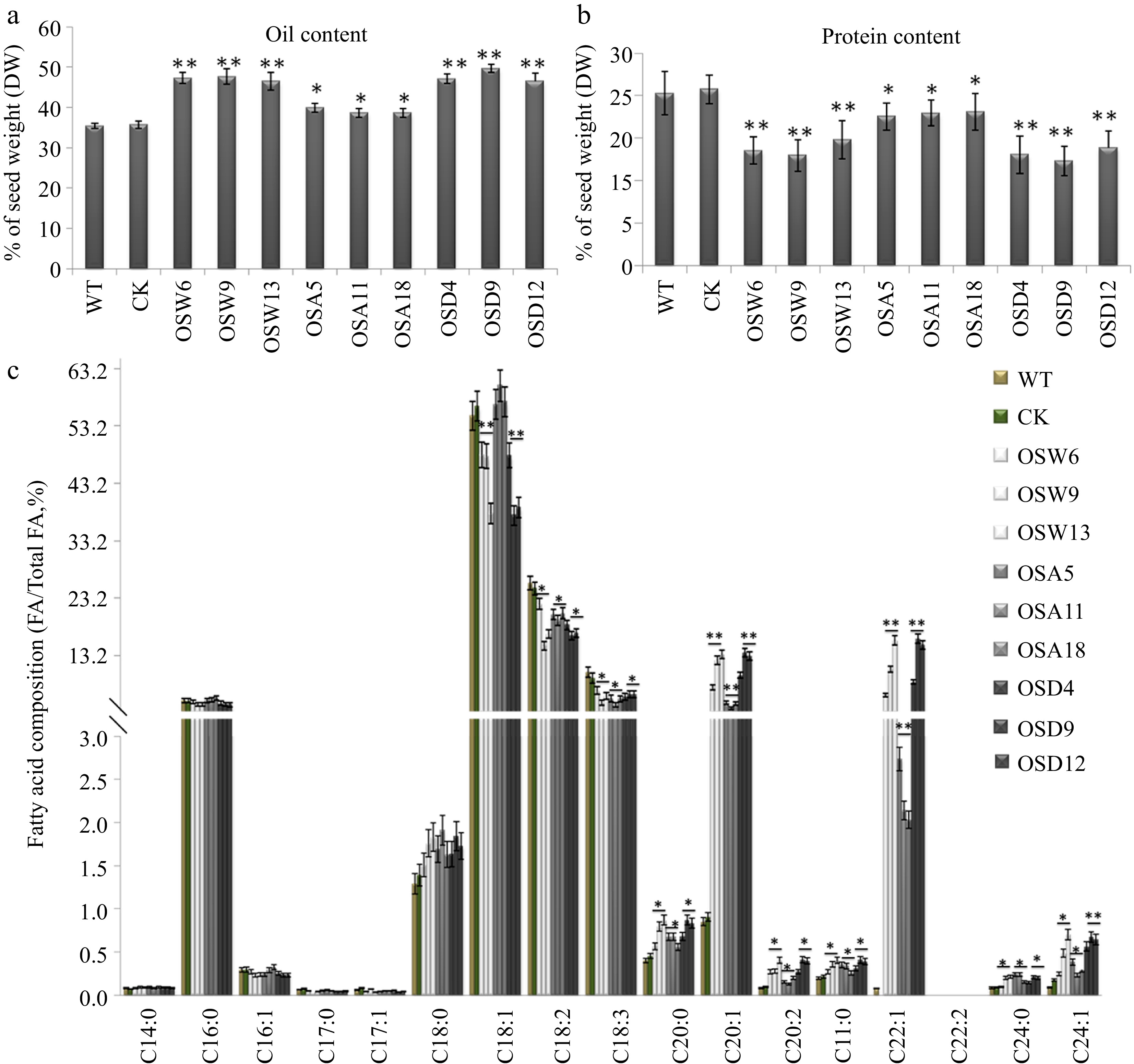
Figure 2.
Assay of oil and protein contents and fatty acid composition in seeds of the BnSnRK2;2, BnSnRK2;2T177A and BnSnRK2;2T177D overexpression transgenic B. napus. Assay of (a) oil content and (b) protein content in seeds of the BnSnRK2;2, BnSnRK2;2T177A and BnSnRK2;2T177D transgenic lines and controls (WT and CK). The seeds of 10 plants in each line (T3 generation) were used for analysis. Means ± SE are shown (Fisher's Least Significant Difference (LSD) test, * p < 0.05, ** p < 0.01). (c) Assay of fatty acid composition in seeds of the BnSnRK2;2, BnSnRK2;2T177A and BnSnRK2;2T177D transgenic lines (T3 generation) and controls. Three biological replicates were performed, and means ± SE are shown (Student's t test, * p < 0.05, ** p < 0.01). WT, wild type; CK, the null transgenic lines; OSW6/9/13, BnSnRK2;2 transgenic lines; OSA5/11/18, BnSnRK2;2T177A transgenic lines; OSD4/9/12, BnSnRK2;2T177D transgenic lines; DAP, day after pollination.
-
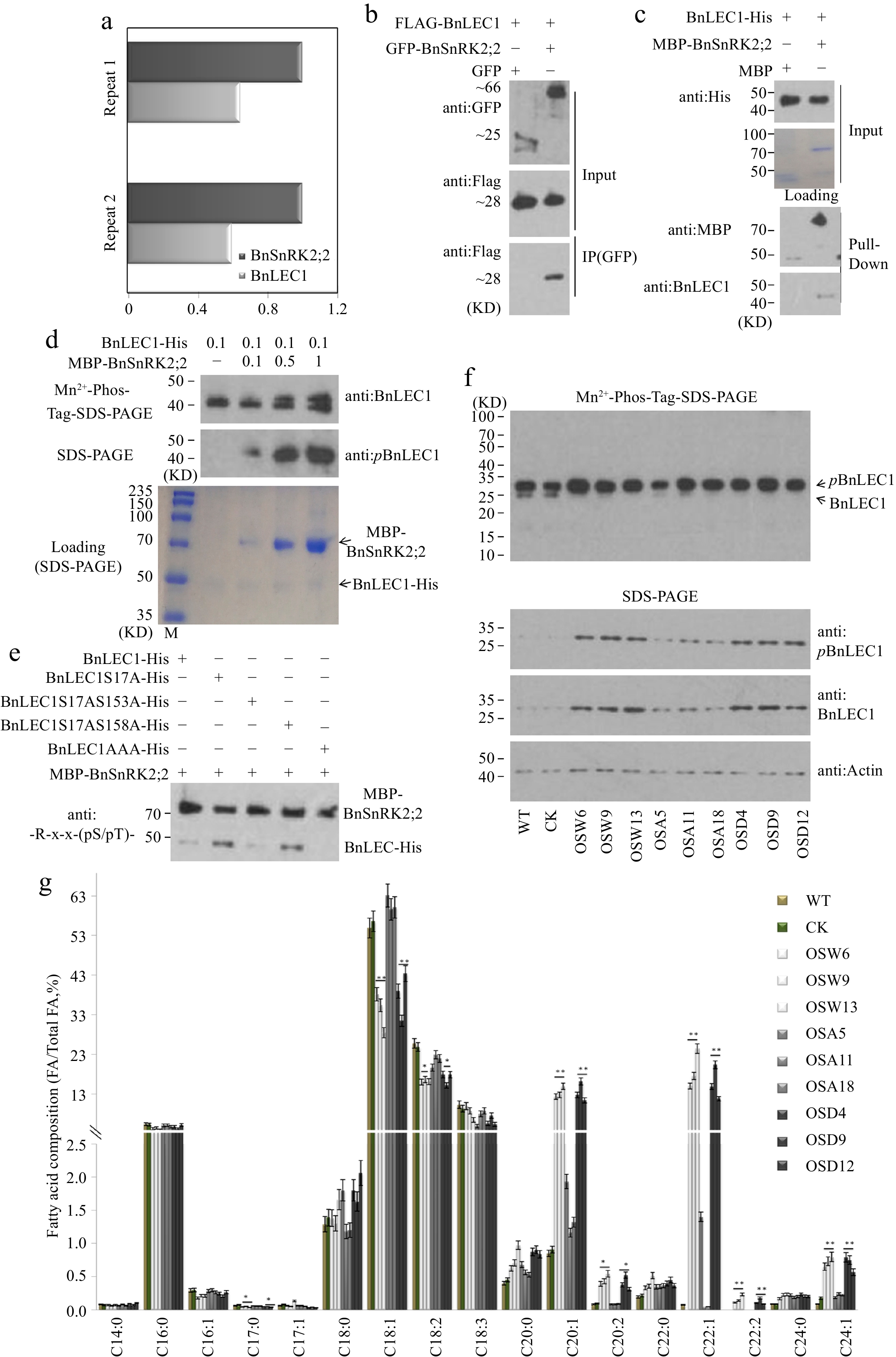
Figure 3.
Assay of BnSnRK2;2 mediated phosphorylation of BnLEC1 in seeds of B. napus. (a) Co-immunoprecipitation (Co-IP) assay of BnSnRK2;2 protein from 25 DAP seeds of B. napus. Proteins were precipitated from cell lysates using anti:BnSnRK2;2 antibody and identified by LC-MS/MS. Control precipitations were performed without antibody. Relative abundance represent the ratio of the number of peptide spectral matches derived from BnLEC1 and BnSnRK2;2. Two biological replicates were performed. (b) Co-IP assay of the interaction of BnSnRK2,2 with BnLEC1 in tobacco (N. benthamiana) leaves co-expressing Flag-BnLEC1 and GFP-BnSnRK2,2 (GFP as the negative control ). The anti-Flag and anti-GFP antibodies were used for immunoblotting analysis. (c) Pull-down assay of BnLEC1-His interacting with MBP-BnSnRK2;2. His-BnLEC1 proteins were incubated with MBP or MBP-BnSnRK2;2 proteins coupled with Amylose Resin. The anti-His and anti-MBP antibodies were used for immunoblotting analysis. (d) In vitro kinase assay was performed using BnLEC1-His and different amount of MBP-BnSnRK2;2 (µg). Immunoblotting analysis using anti:pBnLEC1 antibody was performed to detect the phosphorylation of Ser17 residue in BnLEC1. Proteins were separated by SDS-PAGE (down) or Mn2+-phos-tag gel electrophoresis (up). (e) In vitro kinase assay was performed using five versions of BnLEC1 and MBP-BnSnRK2;2. anti:-R-x-x-(pS/pT)- antibody was used for detecting the phosphorylation of BnLEC1. (f) Protein immunoblotting analysis of phosphorylation of BnLEC1 Ser17 residue in 25 DAP seeds of BnSnRK2;2 transgenic Brassica napus. Proteins were separated by SDS-PAGE (down) or Mn2+-phos-tag gel electrophoresis (up). (g) Assay of fatty acid composition in seeds of BnLEC1, BnLEC1S17A and BnLEC1S17D transgenic lines (T3 generation) and controls (WT and CK). Three biological replicates were performed, and means ± SE are shown (Student's t test, * p < 0.05, ** p < 0.01). WT, wild type; CK, the null transgenic lines; OSW6/9/13, BnSnRK2;2 transgenic lines; OSA5/11/18, BnSnRK2;2T177A transgenic lines; OSD4/9/12, BnSnRK2;2T177D transgenic lines; OLW3/9/14, BnLEC1 transgenic lines; OLA4/13/17, BnLEC1S17A transgenic lines; OLD2/9/15, BnLEC1S17D transgenic lines.
-
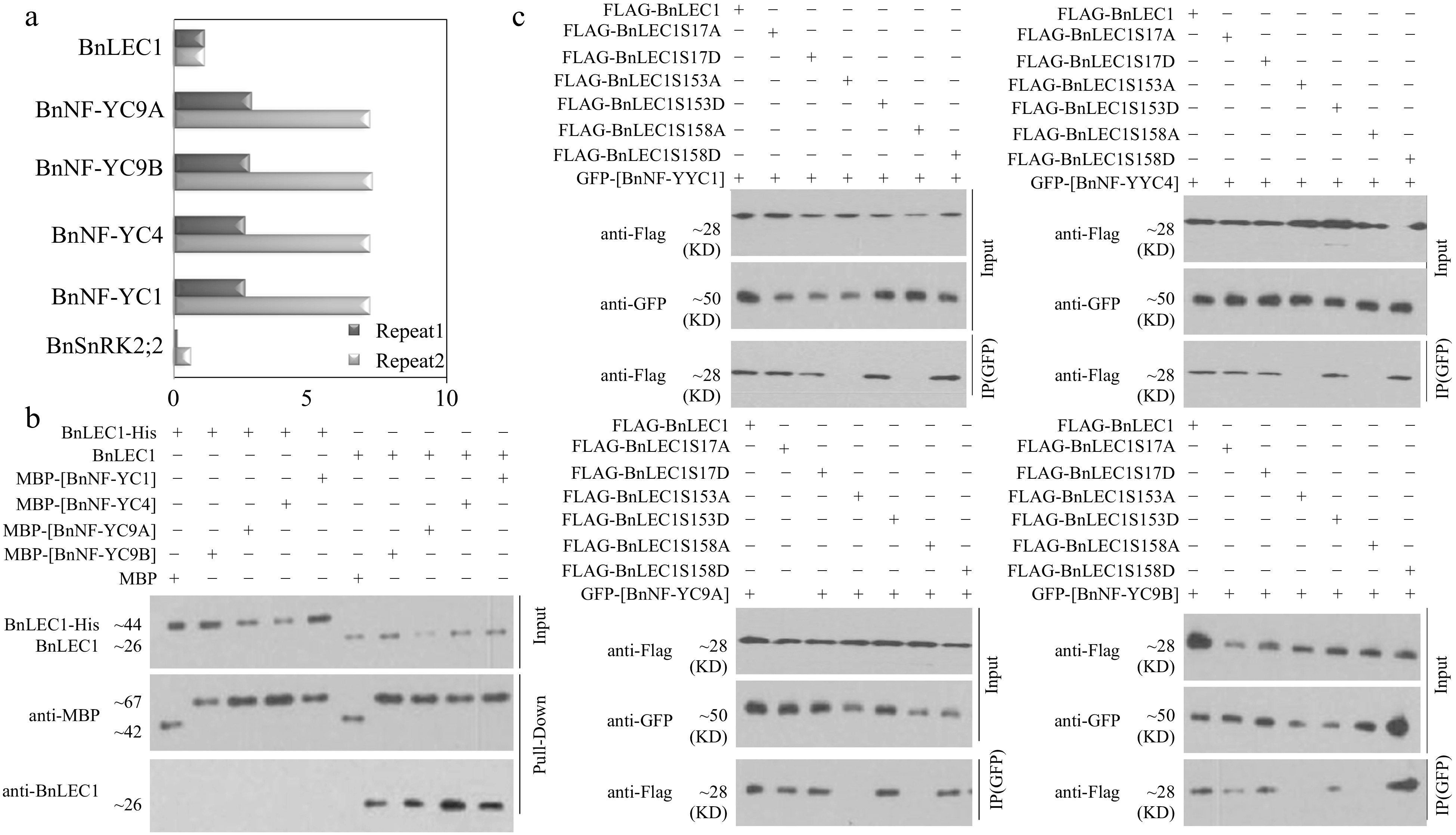
Figure 4.
Assay of BnLEC1 interacting with BnNF-YCs. (a) Co-immunoprecipitation (Co-IP) assay of BnLEC1 protein from 25 DAP seeds of B. napus. Proteins were precipitated from cell lysates using different antibodies and identified by LC-MS/MS. Control precipitations were performed without antibody. Relative abundance represent the ratio of the number of peptide spectral matches derived from the interaction of BnLEC1 with BnNF-YCs and BnSnRK2;2 and BnLEC1 itself. Two biological replicates were performed. (b) Pull-down assay of [BnNF-YCs] and BnLEC1 interaction. His-LEC1 proteins (left) or native LEC1 which was collected by IP from 25 DAP seeds of Brassica napus (right) were incubated with MBP or MBP-[BnNF-YCs] proteins coupled with Amylose Resin. The anti-BnLEC1 or anti-MBP antibody was used for immunoblotting analysis. (c) Co-IP assay of the interaction of BnNF-YCs with BnLEC1 in tobacco (N. benthamiana) leaves coexpressing GFP-[BnNF-YCs] and Flag-BnLEC1. The anti-Flag and anti-GFP antibodies were used for immunoblotting analysis.
-
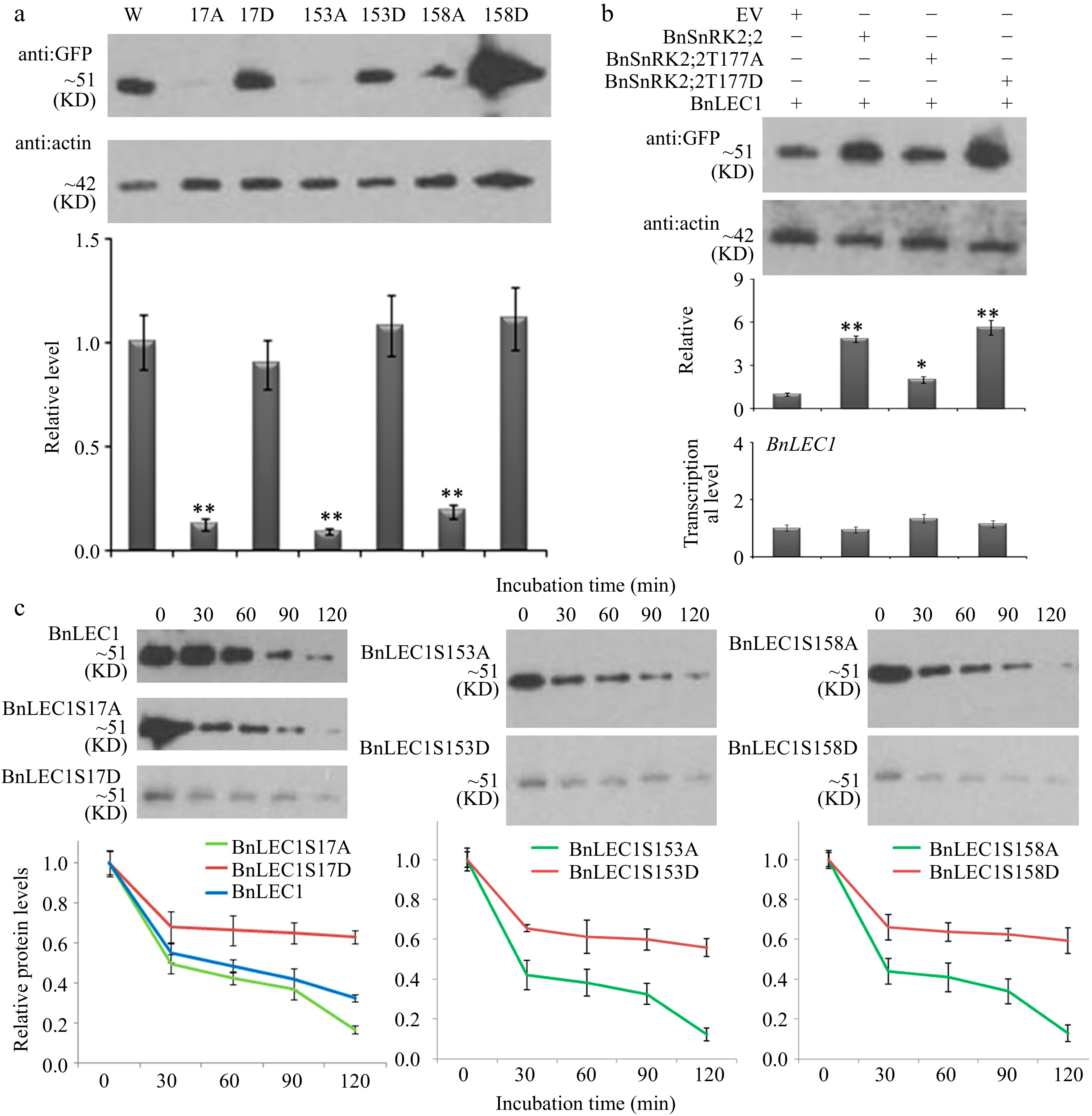
Figure 5.
Assay of BnLEC1 protein stability by its phosphorylation. (a) Protein immunoblotting analysis of the extracts of tobacco (N. benthamiana) leaves expressing NGFP-BnLEC1, NGFP-BnLEC1S17A, NGFP-BnLEC1S17D, NGFP-BnLEC1S153A, NGFP-BnLEC1S153D, NGFP-BnLEC1S158A and NGFP-BnLEC1S158D, respectively. (b) Protein immunoblotting analysis of extracts of tobacco (N. benthamiana) leaves coexpressing NGFP-BnLEC1 and EV (PC2300-NFlag), NFlag-BnSnRK2;2, NFlag-BnSnRK2;2T177A, or NFlag-BnSnRK2;2T177D. (c) Cell-free degradation assay of BnLEC1, BnLEC1S17A, BnLEC1S17D, BnLEC1S153A, BnLEC1S153D, BnLEC1S158A and BnLEC1S158D collected from in vitro kinase reactions containing BnSnRK2;2 and seven versions of BnLEC1, respectively. Equal amounts of BnLEC1, BnLEC1S17A, BnLEC1S17D, BnLEC1S153A, BnLEC1S153D, BnLEC1S158A and BnLEC1S158D were incubated with crude extracts of B. napus leaves supplemented with ATP for time as indicated (in minutes). Immunoblotting assay was performed during cell degradation assays. Three biological replicates were performed. Quantitation of BnLEC1 was achieved using Image J software. Means ± SE are shown (Student's t test, * p < 0.05, ** p < 0.01).
-
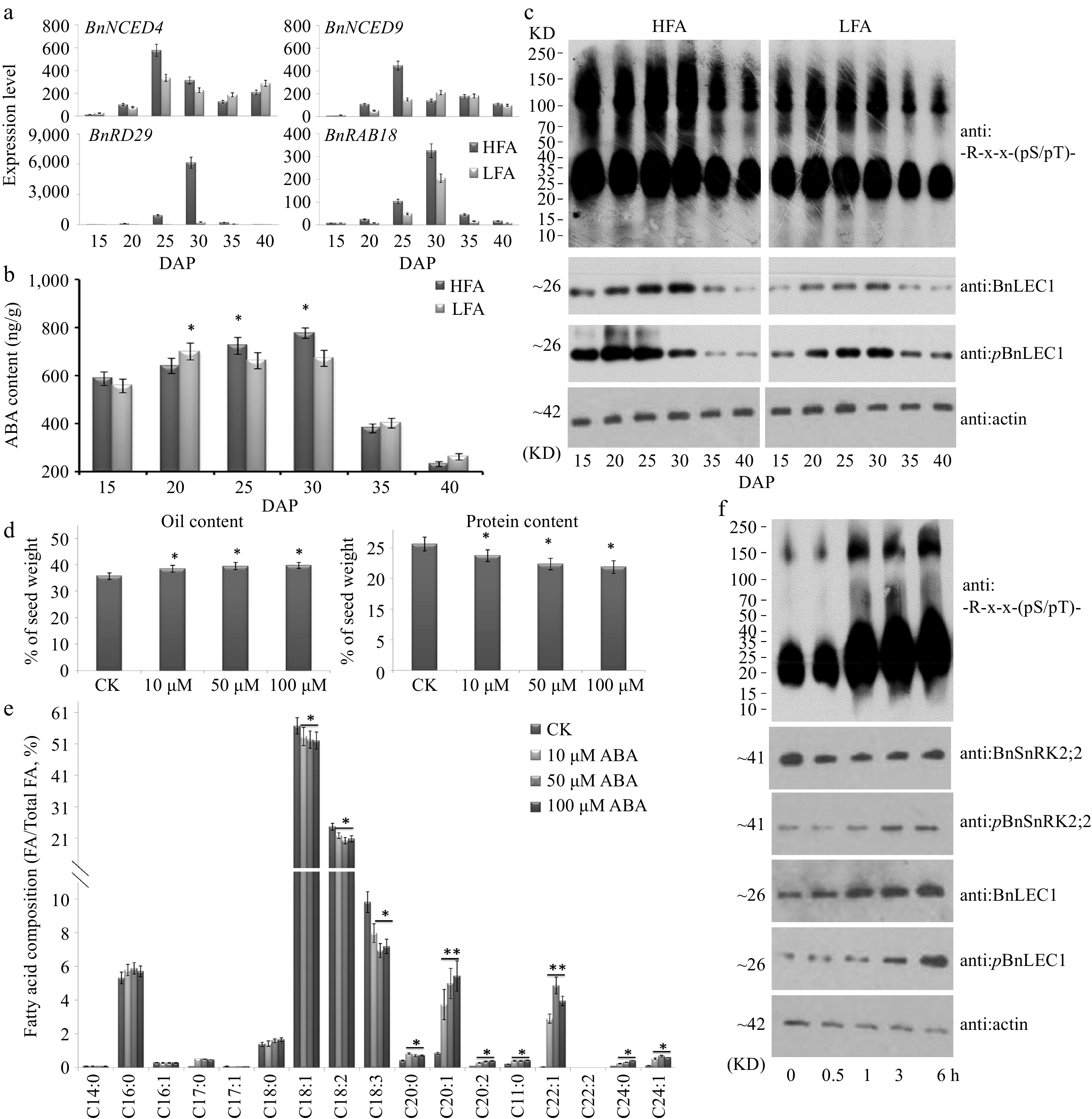
Figure 6.
Assay of abscisic acid (ABA) biosynthesis and physiological function in seed oil accumulation of B. napus. (a) RT-qPCR analysis of time-course expression of ABA biosynthesis genes BnNCED4 and BnNCED9 and ABA-induced maker genes BnRAB18 and BnRD29 in developing seeds of the B. napus varieties HFA and LFA. (b) Quantitation of ABA time-course levels in developing seeds of HFA and LFA. (c) Time-course protein immunoblotting analysis during seed development of HFA and LFA using anti: -R-x-x-(pS/pT)- antibody. Assay of (d) oil and protein contents and (e) fatty acid composition in seeds after ABA treatment. Fifty µM ABA and water (CK) was injected into pod cavities of B. napus (cv. Westar) and sucked out half an hour later with injector from 20 DAP to 35 DAP every 2 d. Then mature seeds were used for analyzing oil and protein contents and fatty acid composition. (f) Protein immunoblotting analysis were performed to detect the influence of exogenous ABA in 25 DAP seeds using antibody of anti:BnLEC1, anti:pBnLEC1, anti:BnSnRK2;2, anti:pBnSnRK2;2 and anti: -R-x-x-(pS/pT)-. Three biological replicates were performed, and means ± SE are shown (Student's t test, * p < 0.05, ** p < 0.01). HFA, a Brassica napus line with high seed oil content; LFA, a Brassica napus line with low seed oil content; DAP, day after pollination.
-
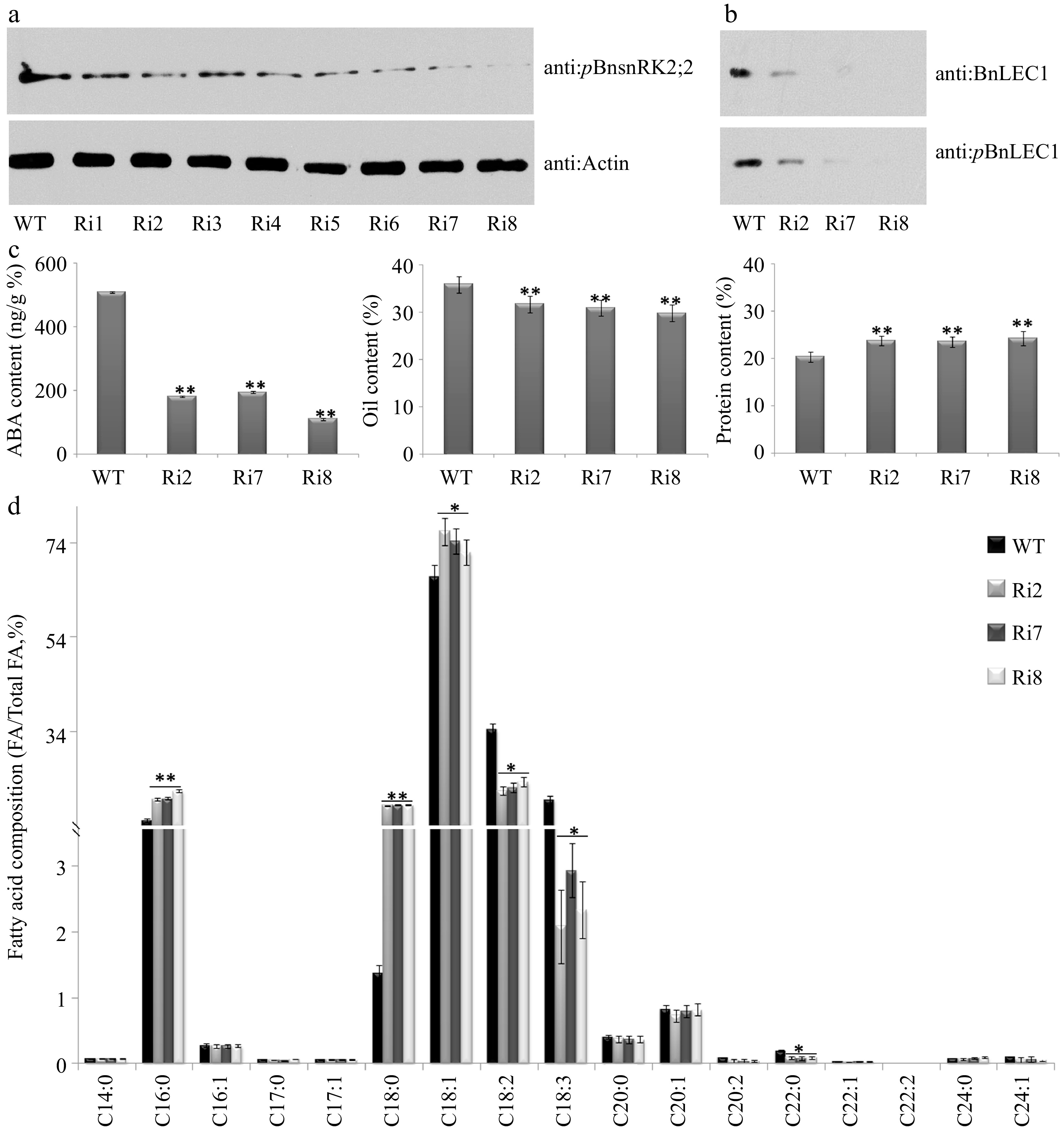
Figure 7.
Assay of oil and protein contents and fatty acid composition in seeds of the BnABA2 RNAi transgenic B. napus. (a) Protein immunoblotting analysis of phosphorylation of BnSnRK2;2 in 25 DAP seeds of BnABA2 RNAi transgenic Brassica napus. Proteins were separated by SDS-PAGE. The anti-pBnSnRK2;2 or anti-actin (control) antibody was used for immunoblotting analysis. (b) Protein immunoblotting analysis of phosphorylation of BnLEC1 at Ser17 residue in 25 DAP seeds of BnABA2 RNAi transgenic B. napus. Proteins were separated by SDS-PAGE as in (a). The anti-BnLEC1 or anti-pBnLEC1 antibody was used for immunoblotting analysis. (c) Assay of ABA content, oil content and protein content in 25 DAP seeds of the BnABA2 RNAi transgenic lines and wild type. The seeds of 10 plants in each line (T2 generation) were used for analysis. Three biological replicates were performed, and means ± SE are shown (Fisher's Least Significant Difference (LSD) test, * p < 0.05, ** p < 0.01). (d) Assay of fatty acid composition in seeds of the BnABA2 RNAi transgenic lines (T2 generation) and WT. Three biological replicates were performed, and means ± SE are shown (Student's t test, * p < 0.05, ** p < 0.01). WT, wild type; Ri, BnABA2 RNAi transgenic lines; DAP, day after pollination.
-
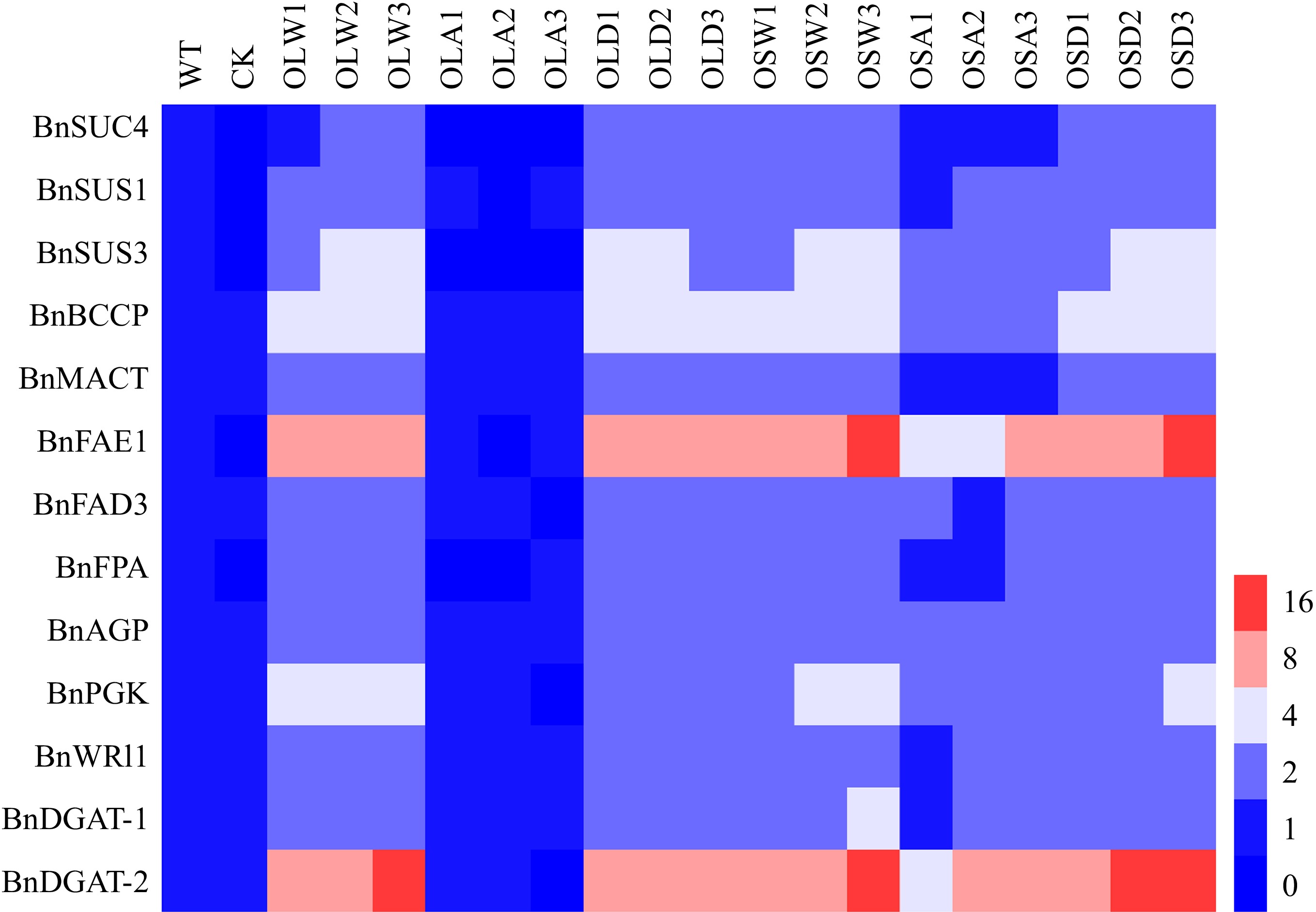
Figure 8.
BnSnRK2;2 mediated phosphorylation of BnLEC1 affects the expressions of the genes related to sucrose photo-assimilation, glycolysis, and fatty acid (FA) biosynthesis and modification. RT-qPCR analysis of the expressions of BnSUS1, BnSUS3, BnSUC4, BnAGP, BnPGK, BnFPA, BnBCCP, BnMACT, BnDGAT1, BnDGAT2, BnFAE1 and BnFAD3 in 25 DAP seeds of the BnLEC1 and BnLEC1S17A, BnLEC1S17D, BnSnRK2;2, BnSnRK2;2T177A and BnSnRK2;2T177D overexpression transgenic lines (T3 generation) and controls (WT and CK). WT, wild type; CK, the null transgenic lines; OLW3/9/14, BnLEC1 transgenic lines; OLA4/13/17, BnLEC1S17A transgenic lines; OLD2/9/15, BnLEC1S17D transgenic lines; OSW6/9/13, BnSnRK2;2 transgenic lines; OSA5/11/18, BnSnRK2;2T177A transgenic lines; OSD4/9/12, BnSnRK2;2T177D transgenic lines.
-
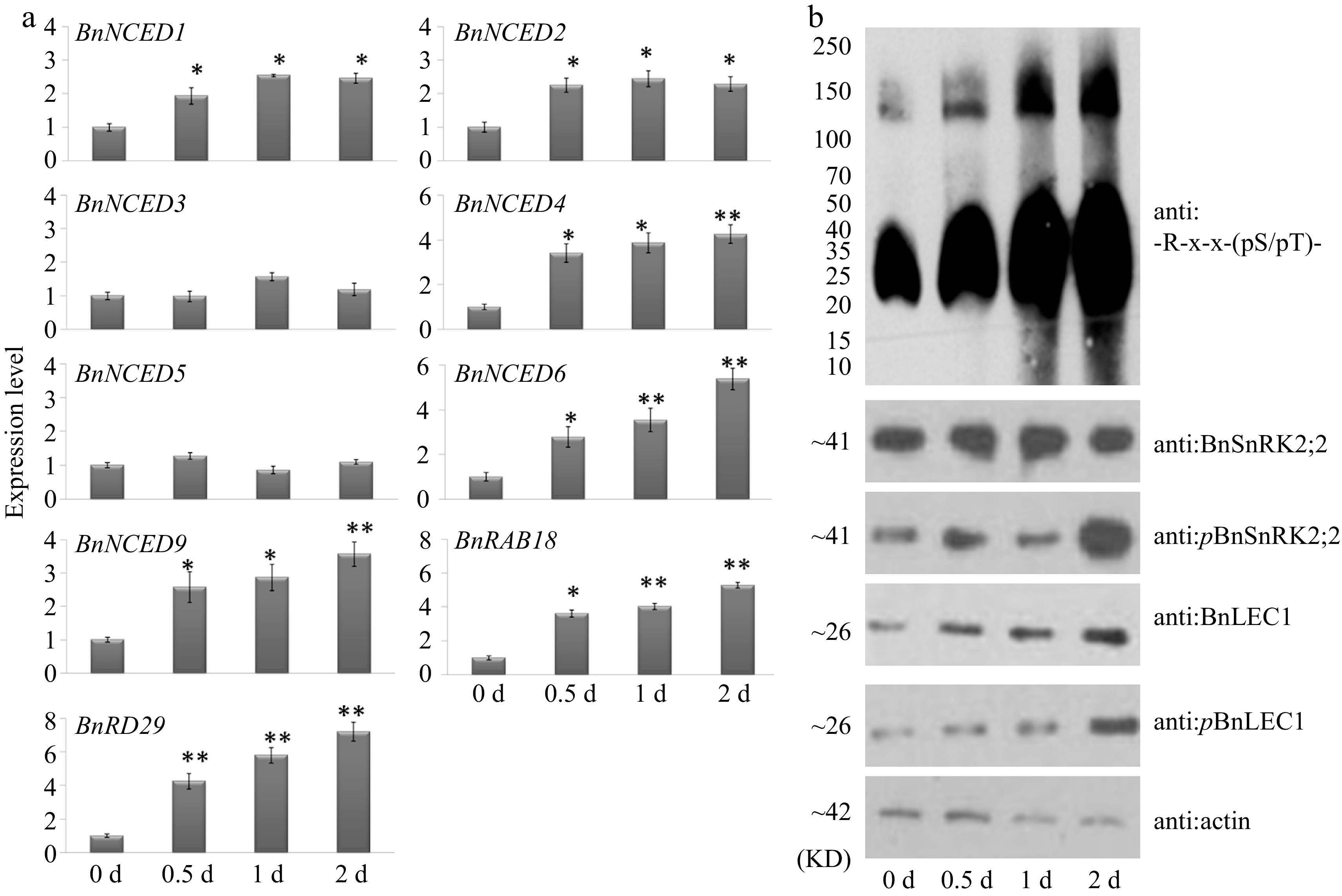
Figure 9.
Glucose promotes expressions of ABA synthesis genes and phosphorylation of BnLEC1 by BnSnRK2;2 in seeds of B. napus. (a) RT-qPCR analysis of the expressions of ABA biosynthesis genes in 25 DAP seeds after glucose treatment for 0–2 d. Three biological replicates were performed, and means ± SE are shown (Student's t test, * p < 0.05, ** p < 0.01). (b) Protein immunoblotting analysis were performed to detect the influence of exogenous glucose in 25 DAP seeds using antibodies of anti:BnLEC1, anti:pBnLEC1, anti:BnSnRK2;2, anti:pBnSnRK2;2 and anti:-R-x-x-(pS/pT)-. Three biological replicates were performed.
-
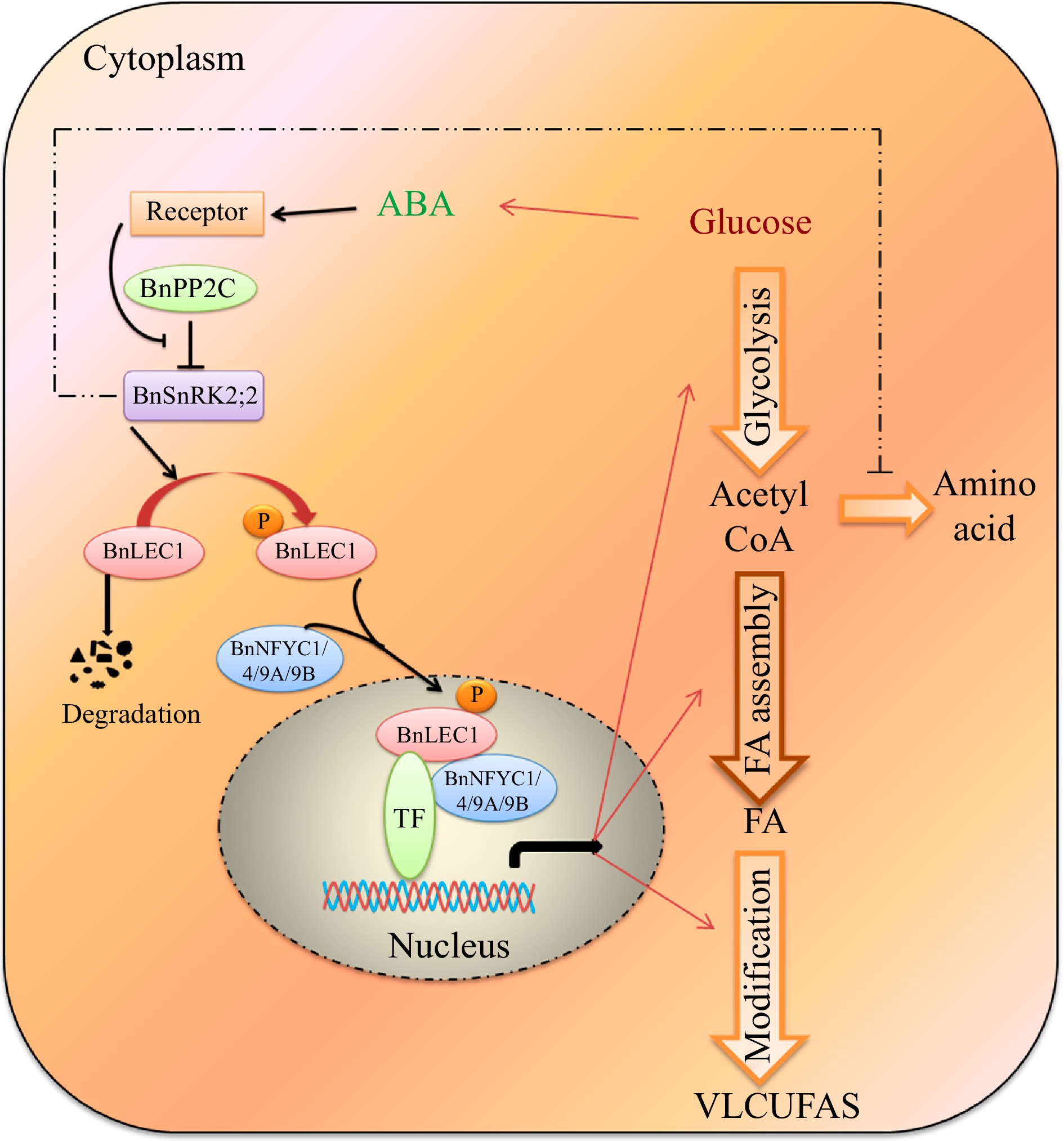
Figure 10.
A proposed model for ABA-dependent BnSNRK2;2 BnLEC1-[BnNF-YCs] signaling cascade promote flow of carbon from glucose to lipid biosynthesis in response to intracellular sugar state. During seed development of Brassica napus, glucose may promote the ABA biosynthesis, and ABA signaling enhances the phosphorylation of BnLEC1 by BnSnRK2;2. Then, the phosphorylated BnLEC1 forms heterodimer with BnNF-YC1/4/9A/9B in the cytoplasm, and is translocated into the cell nucleus, where the BnLEC1 complex activates the expressions of the genes related sucrose photo-assimilation, glycolysis, and FA biosynthetic and modification required for lipid biosynthesis and accumulation in seeds. Thus, ABA-BnPP2C-BnSnRK2;2-BnLEC1-[BnNF-YC1/4/9A/9B] cascade promotes flow of carbon from glucose to lipid biosynthesis in response to intracellular sugar state. On the other hand, BnSnRK2;2 might suppress protein synthesis by inhibiting the flux through the tricarboxylic acid cycle. The fine and thick lines represent transcriptional level and post-translational regulation, respectively. The imaginary line represents the regulation need to be further verified in seeds of B. napus.
Figures
(10)
Tables
(0)