-
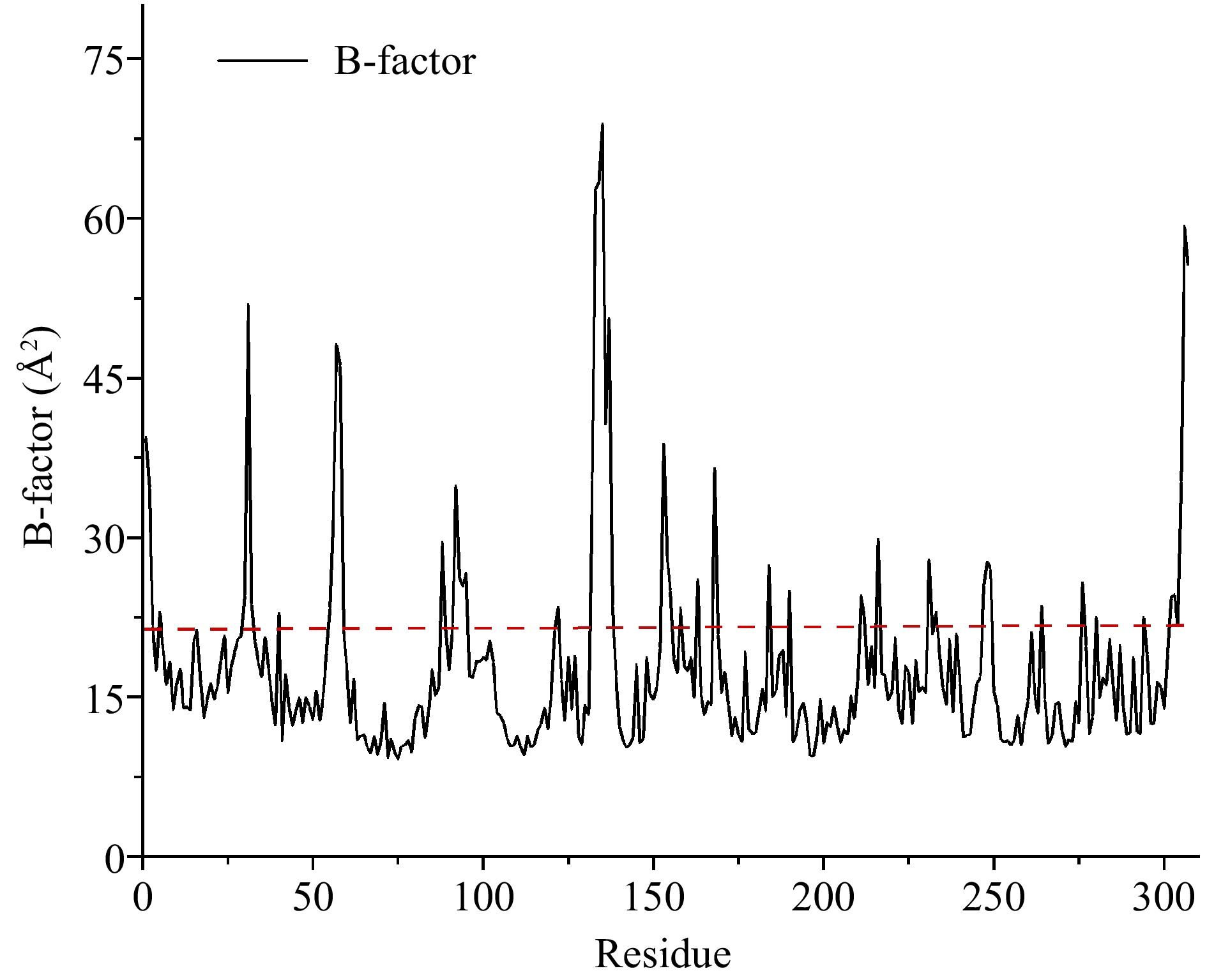
Figure 1.
Average B-factor values of carboxypeptidase A. Flexible residues were located above the dotted line.
-
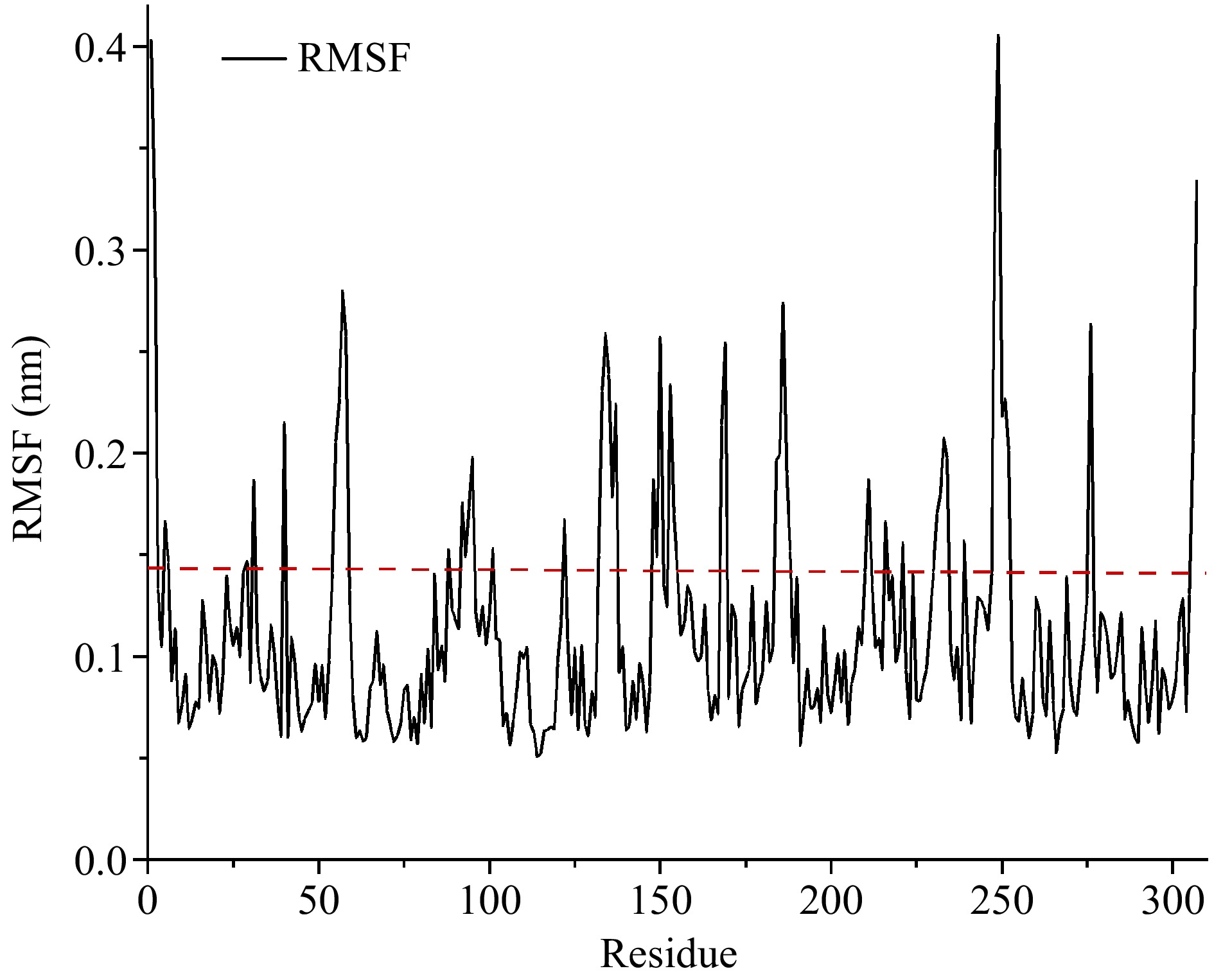
Figure 2.
The RMSF values of carboxypeptidase A. Flexible residues were located above the dotted line.
-

Figure 3.
The comparison of RMSD values between wild-type and mutant carboxypeptidase A. (a) WT and D93C/F96C. (b) WT and K153C/S251C.
-
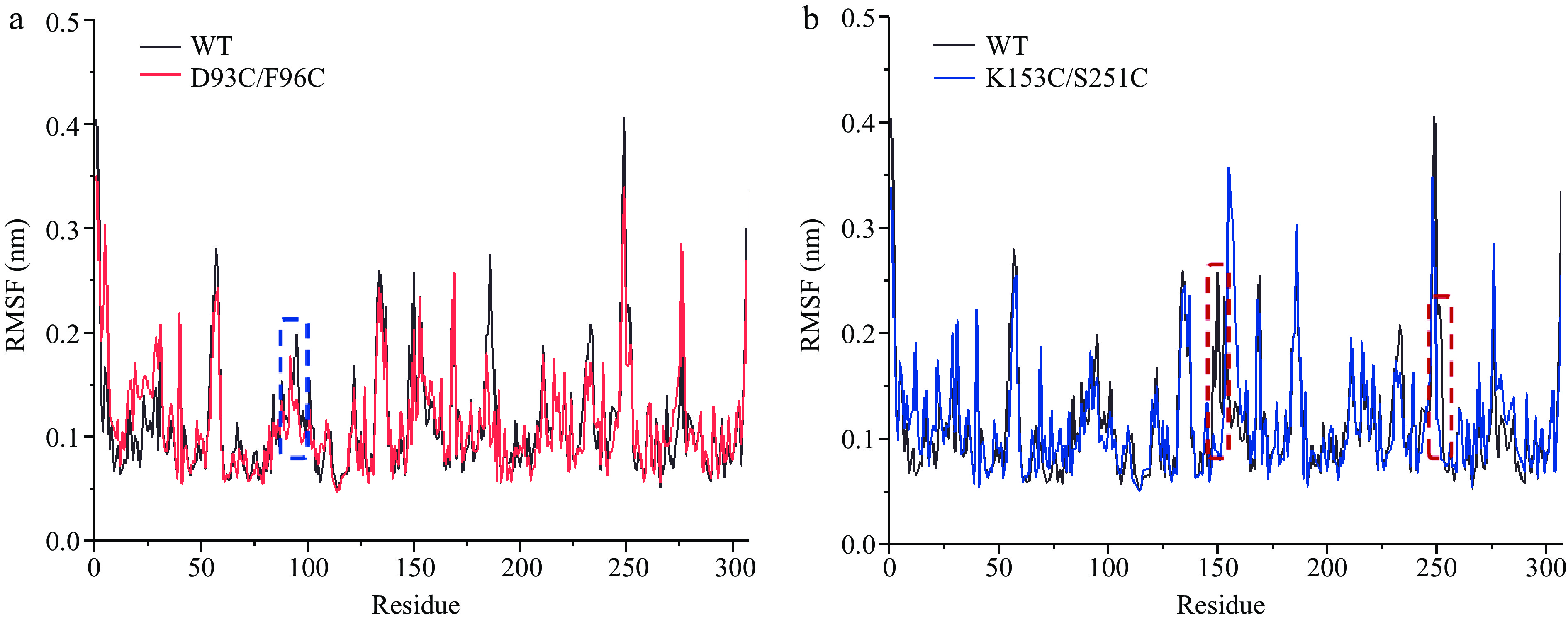
Figure 4.
The comparison of RMSF values between wild-type and mutant carboxypeptidase A. (a) WT and D93C/F96C. (b) WT and K153C/S251C. The mutant regions are circled in dotted wireframes.
-
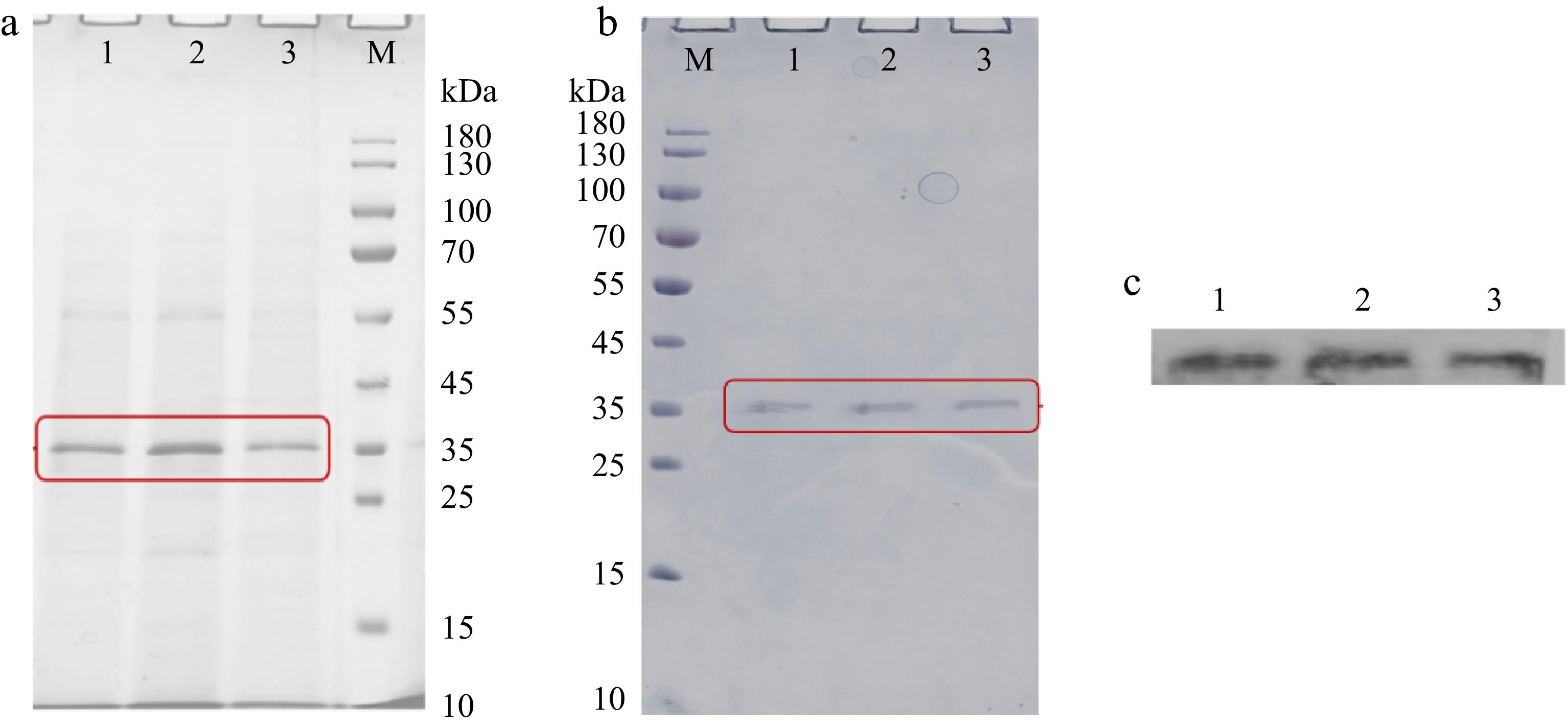
Figure 5.
The SDS-PAGE analysis of (a) fermentation supernatants and (b) purified components expressed by P. pastoris. (c) The western blot analysis of purified components expressed by P. pastoris. M, protein marker (10−180 kDa). 1, WT. 2, D93C/F96C. 3, K153C/S251C. Each sample was prepared by boiling for 5 min and loaded at 20 μL per lane.
-
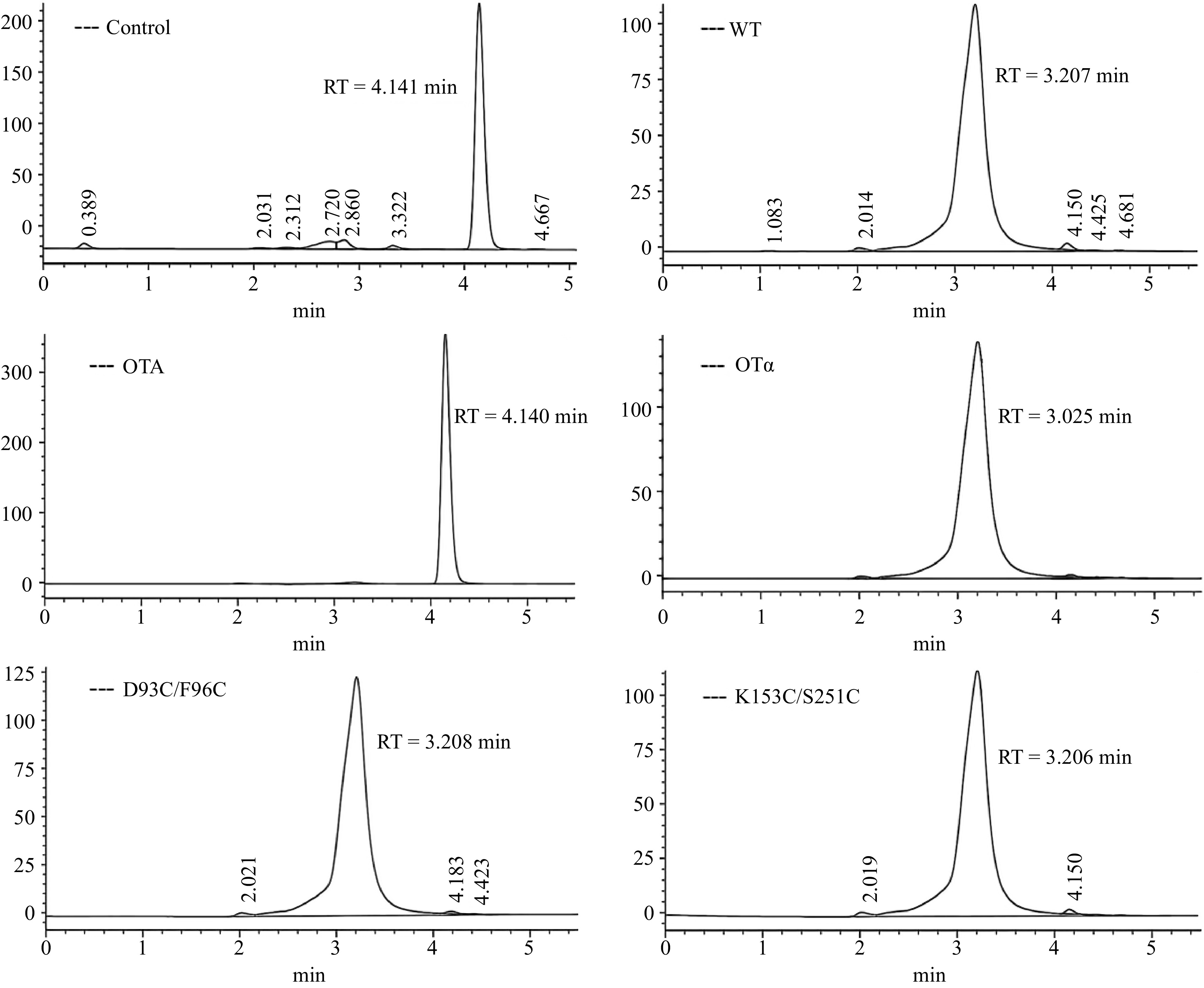
Figure 6.
HPLC of OTA degradation by recombinant CPA and its mutants.
-
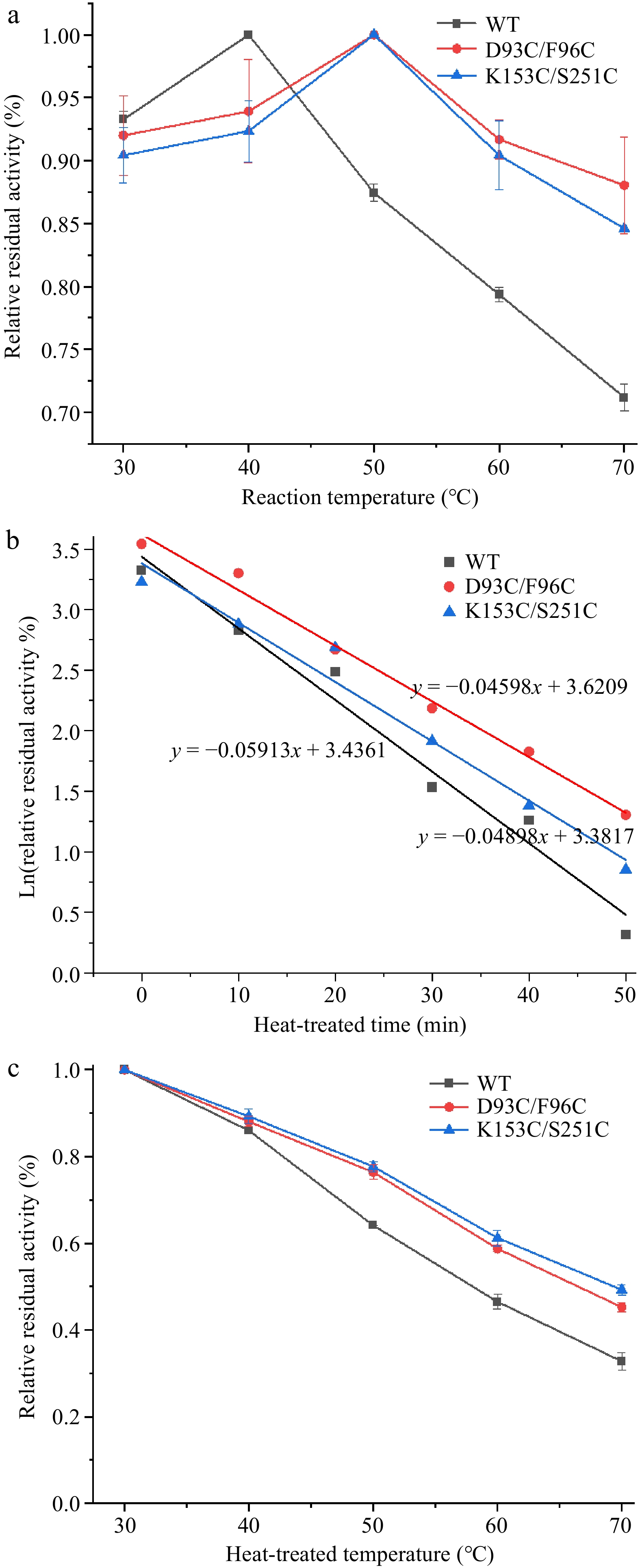
Figure 7.
Thermal stability of wild-type and mutant carboxypeptidase A. (a) The optimum temperature of wild-type and mutant carboxypeptidase A. (b) The half-life of wild-type and mutant carboxypeptidase A. (c) Half inactivation temperature of wild-type and mutant carboxypeptidase A.
-

Figure 8.
The contents of secondary structures (α-helix, β-strand, β-turn, and coil) in CPA and its mutants.
-
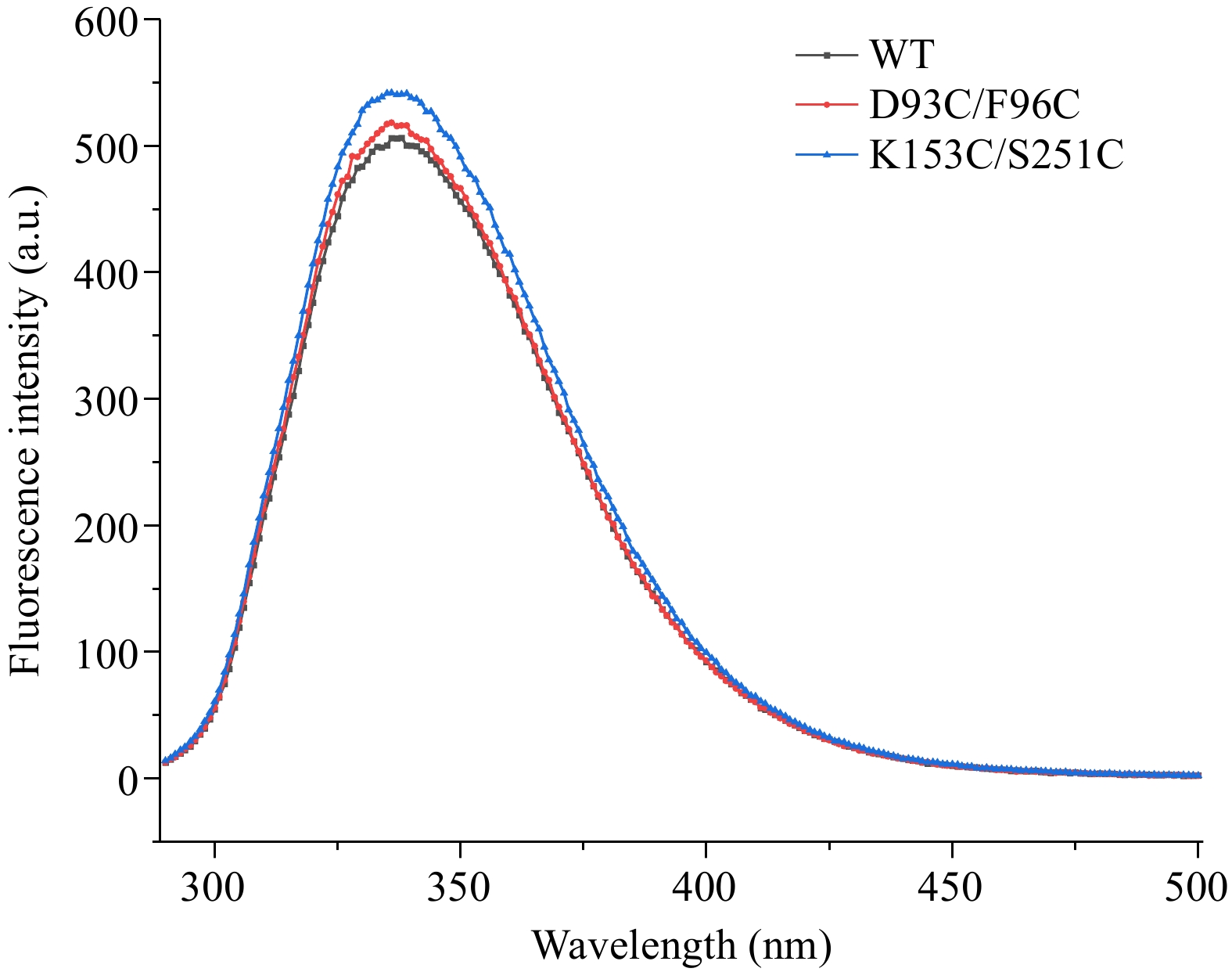
Figure 9.
Intrinsic fluorescence spectra of wild-type and mutant carboxypeptidase A.
-
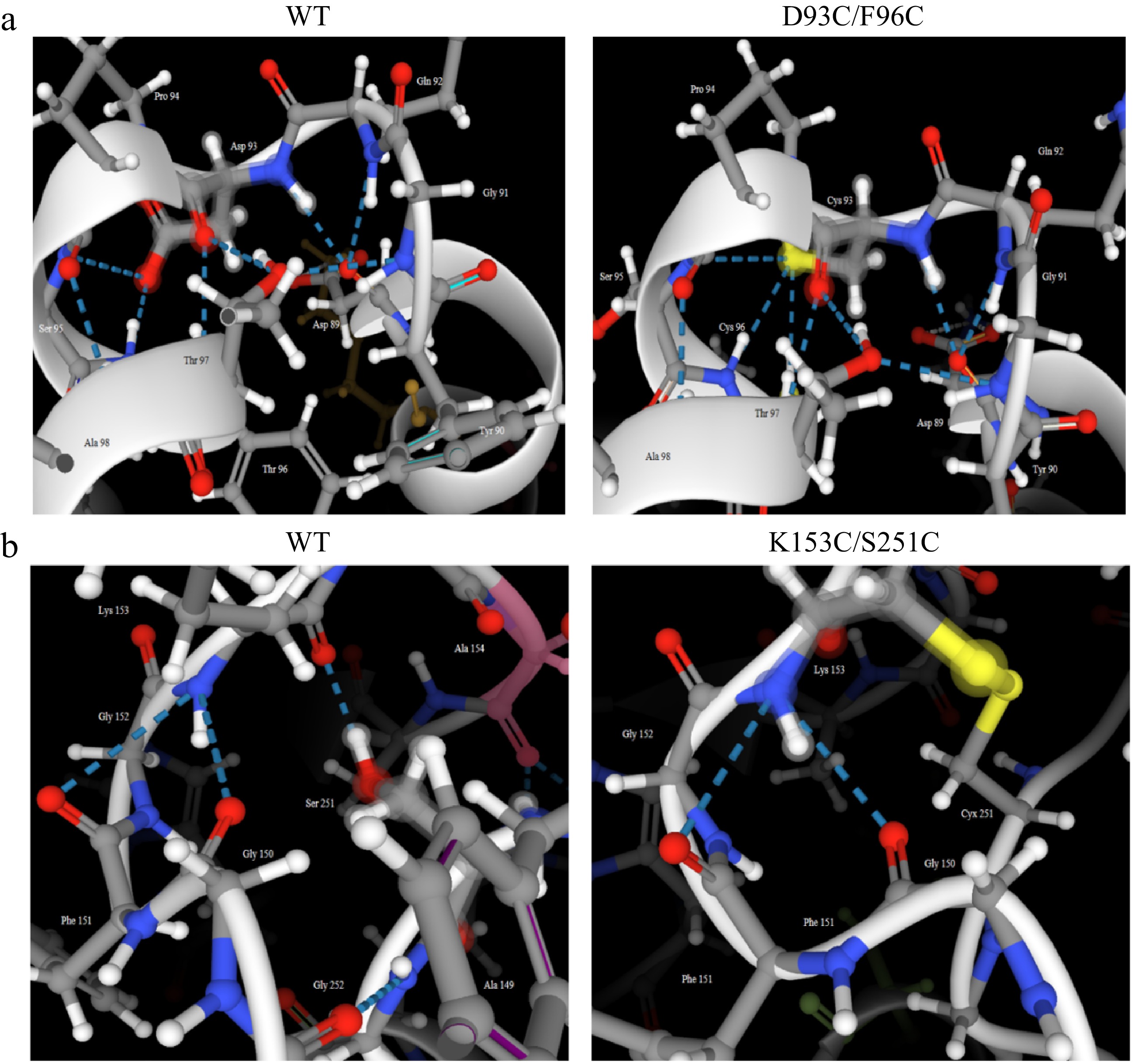
Figure 10.
Comparison of the number of hydrogen bonds in mutant regions between wild-type and its mutant carboxypeptidase A. (a) WT and D93C/F96C. (b) WT and K153C/S251C.
-
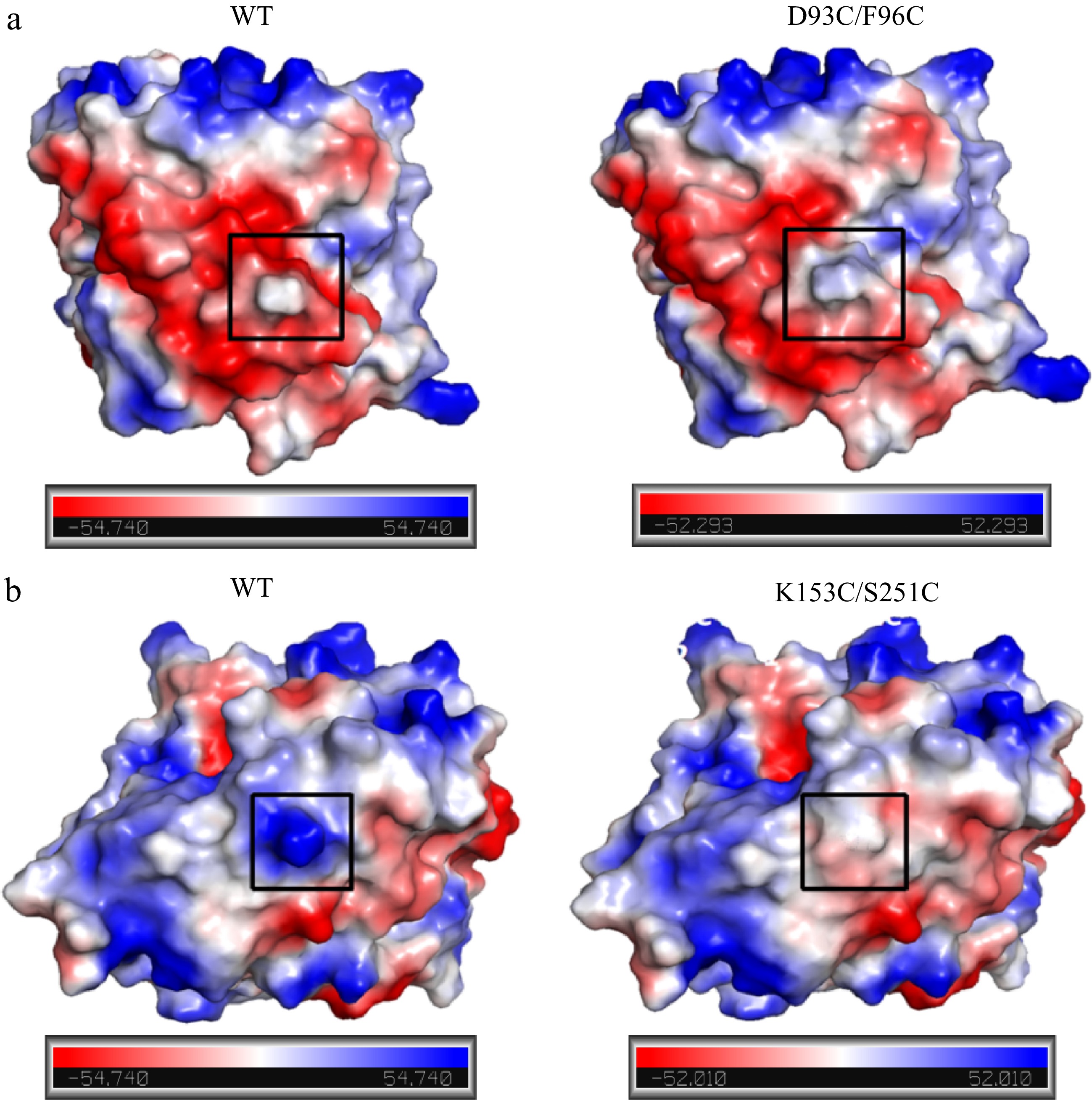
Figure 11.
Comparison of surface charge distribution in mutant regions between wild-type and mutant carboxypeptidase A. (a) WT and D93C/F96C; (b) WT and K153C/S251C. The mutation regions were circled in black wireframes.
-
Enzyme Km (μM) Vmax (μM·min−1) Kcat/Km (μM−1·s−1) WT 0.277 ± 0.012 1.833 ± 0.014 4.549 × 10−3 D93C/F96C 0.271 ± 0.021 2.569 ± 0.036 6.512 × 10−3 K153C/S251C 0.268 ± 0.043 1.641 ± 0.047 4.209 × 10−3 Table 1.
Kinetic parameters of wild-type and mutant carboxypeptidase A.
-
Enzyme A1 A2 Disulfide bond WT 0.456 ± 0.009 0.342 ± 0.010 0 D93C/F96C 1.434 ± 0.035 0.367 ± 0.018 1 K153C/S251C 1.549 ± 0.050 0.464 ± 0.023 1 Table 2.
Comparison of disulfide bond number between wild-type and mutant carboxypeptidase A.
Figures
(11)
Tables
(2)