-
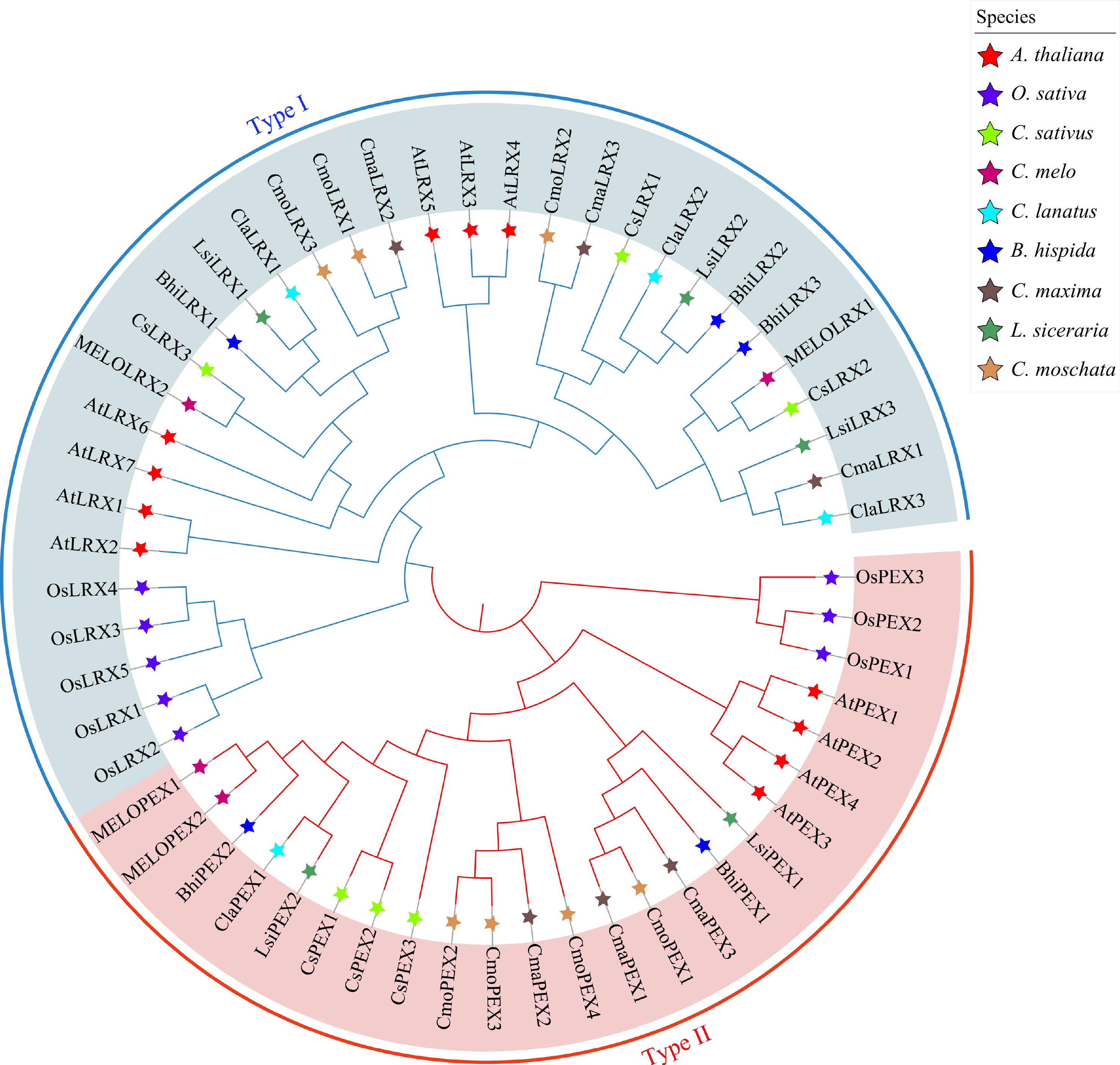
Figure 1.
Phylogenetic tree of the LRX proteins from Arabidopsis, rice, and seven cucurbit species. Red star, purple star, light green star, deep red star, sky blue star, deep blue star, deep brown star, deep green star and light brown star represent Arabidopsis thaliana (A. thaliana), rice (O. sativa), cucumber (C. sativus ), melon (C. melon), watermelon (C. lanatus), wax gourd (B. hispida), pumpkin (C. maxima), bottle gourd (L. siceraria) and moschata pumpkin (C. moschata), respectively.
-

Figure 2.
Phylogenetic clustering, conserved motifs, conserved domains and gene structures of seven cucurbits LRX genes. Left one: The phylogenetic tree of LRX genes. Light green and brownish red represent LRX and PEX subfamilies, respectively. Left two: the conserved motif of LRX genes, different colors represent different motifs; right one: LRX gene structure analysis, green and blue represent the UTR region and the CDS region, respectively; right two: the conserved domain of LRX genes, different colors represent different domains.
-
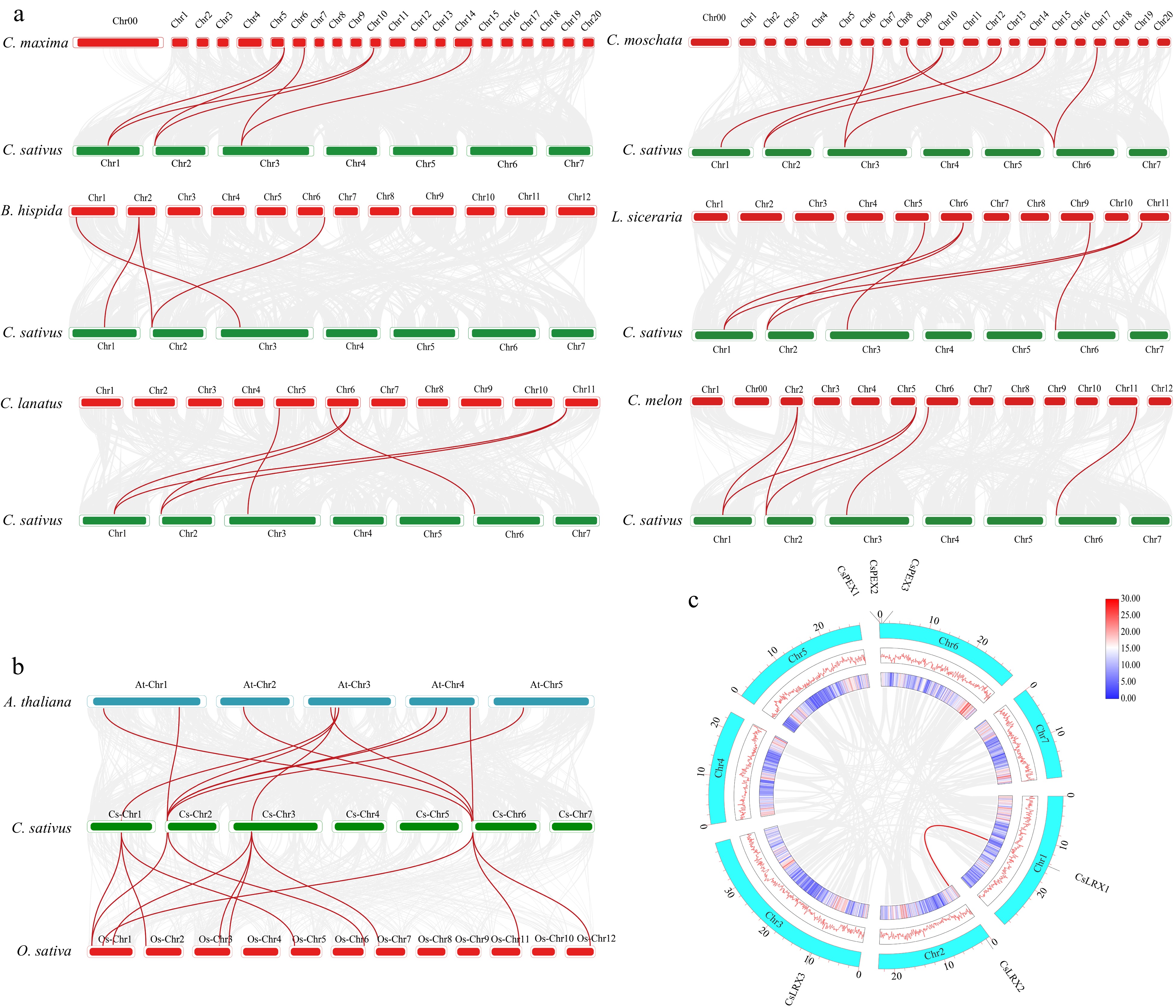
Figure 3.
Synteny analysis of LRX genes. (a) Synteny analysis of LRX genes between cucumber and six other cucurbits. (b) Synteny analysis of LRX genes in cucumber, Arabidopsis thaliana and rice (O. sativa). (c) Colinearity analysis of CsLRX genes in cucumber species. The red and blue lines represent gene pairs with collinearity.
-
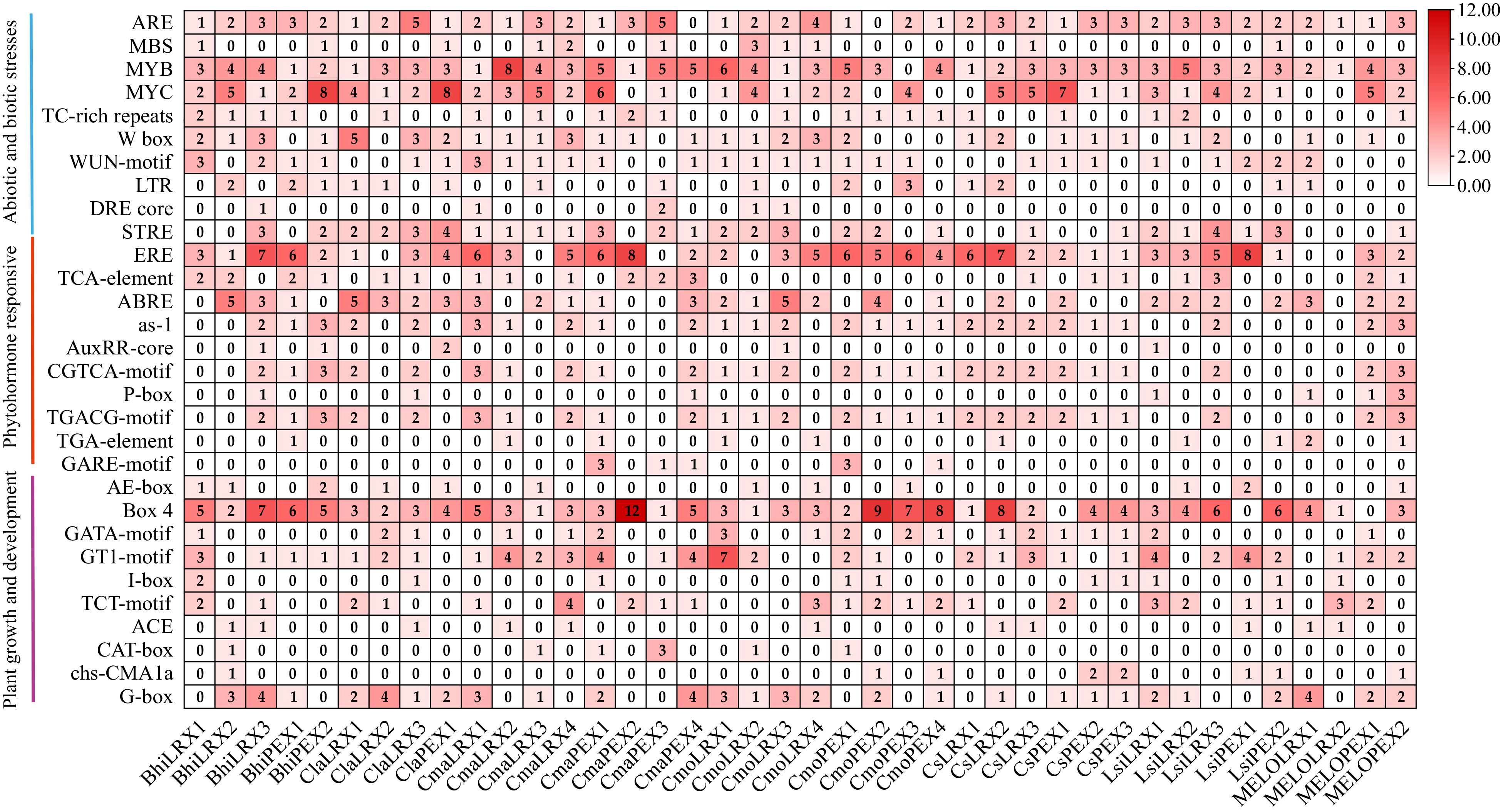
Figure 4.
Cis-acting elements in the promotors of LRX genes in seven cucurbit species. The colors indicate the different cis-elements numbers. Values indicate the statistical number of cis elements.
-
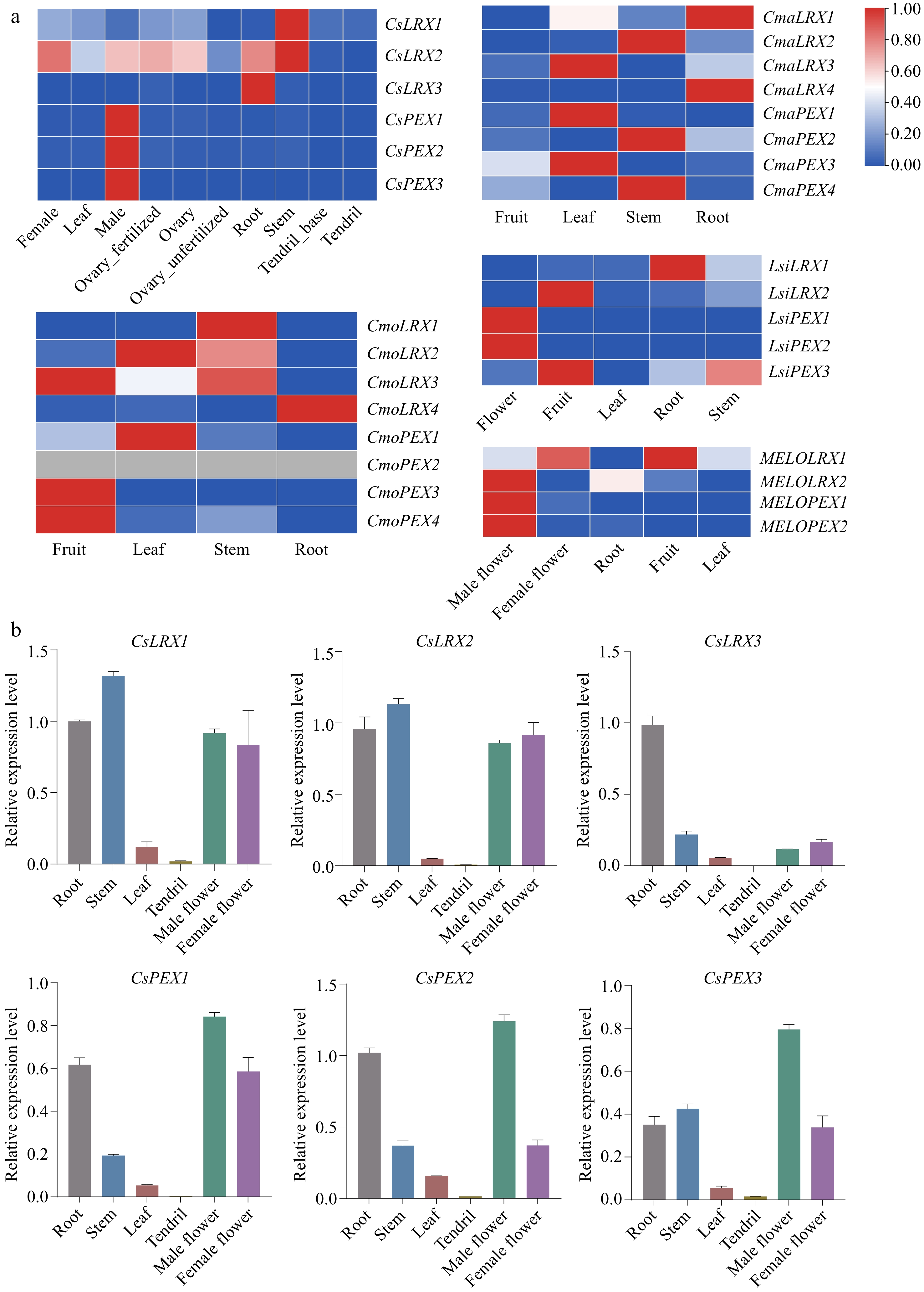
Figure 5.
The expression pattern of LRX genes in different Cucurbitaceae plant tissues. (a) The expression of public RNA-seq data of LRX genes in different tissues of Cucurbitaceae. (b) qRT-PCR analysis of CsLRX genes in different tissues of cucumber. Values are means ± SD of three biological replicates.
-
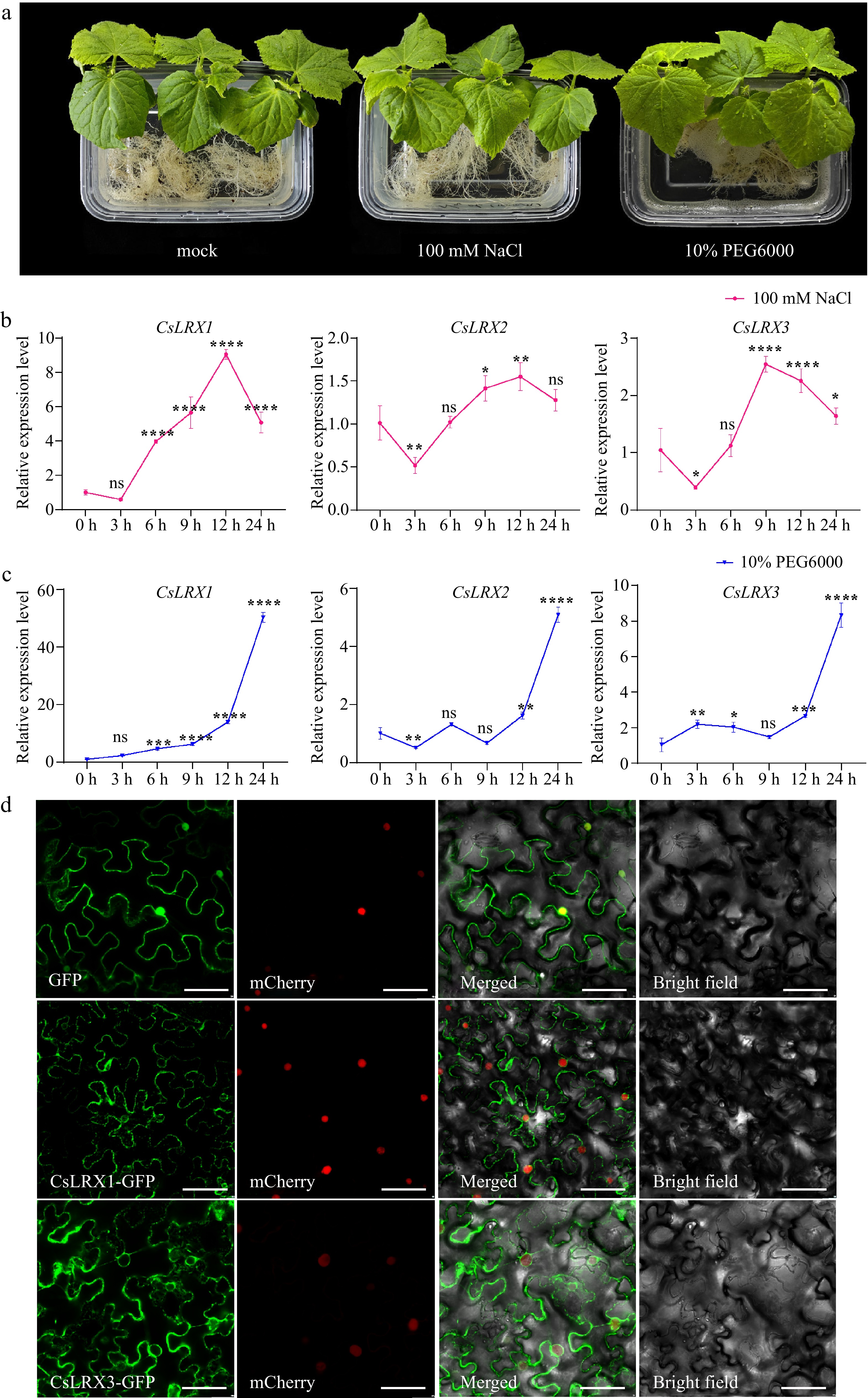
Figure 6.
Expression analysis of CsLRX genes in cucumber. (a) Cucumber roots were treated with 100 mM NaCl and 10% PEG6000, and water was used as a control (mock). (b)−(c) The CsLRX genes in cucumber roots treated with 100 mM NaCl and 10% PEG6000 for different time was analyzed by qRT-PCR. Values are means ± SD of three biological replicates. Significant differences between 0 h and other time points are indicated by asterisks (ns: no significance, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, Student's t test). (d) Subcellular localization of CsLRX1 and CsLRX3. The empty vector was used as a control. These indicated structures were transiently expressed in N. benthamiana leaves. Bar, 50 μm.
-
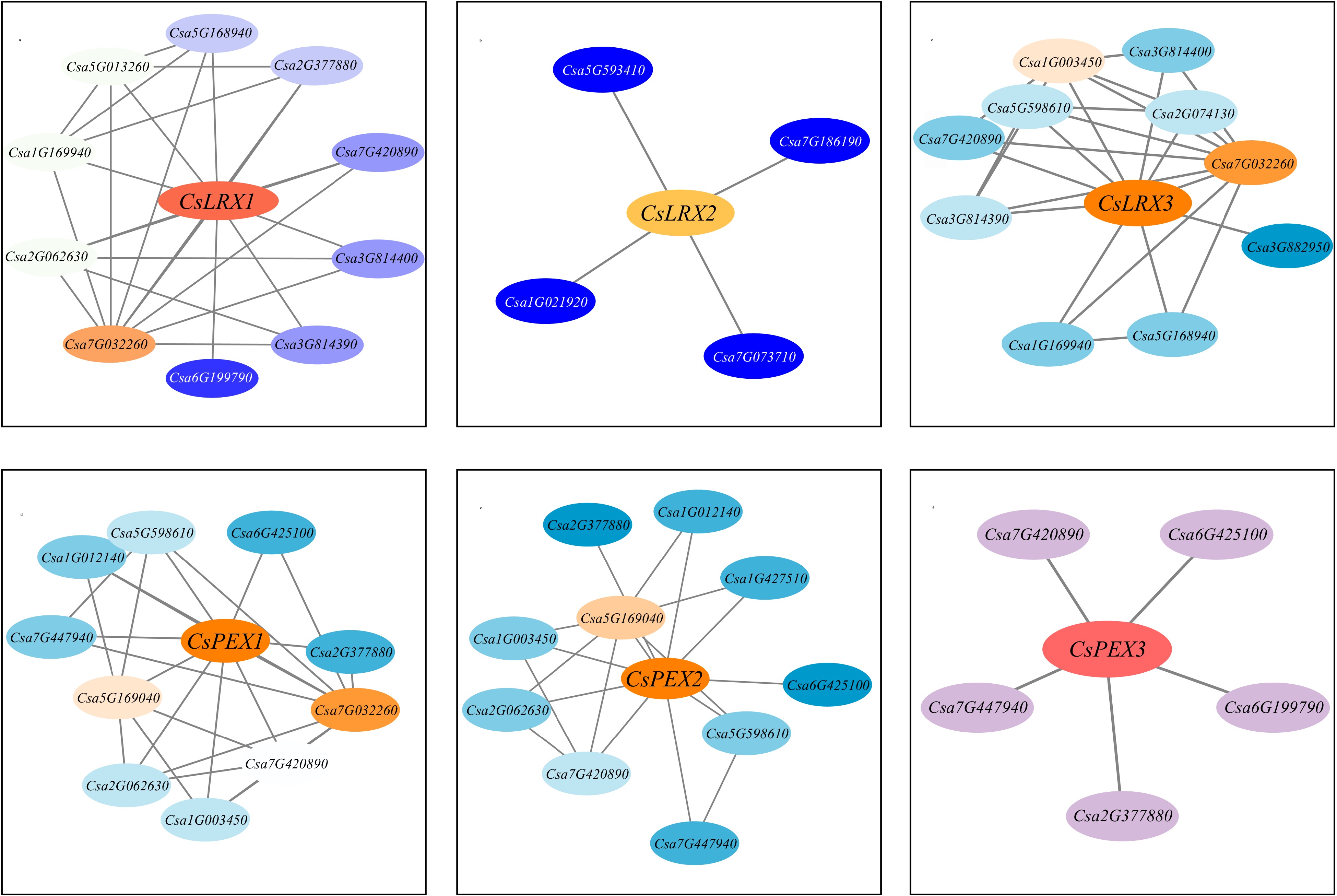
Figure 7.
Interaction network prediction of CsLRX protein in cucumber.
-
Gene name Gene ID Gene position CDS (bp) AA MW (Da) PI Arabidopsis homology Start End (+/−) CsLRX1 Csa1G383520.1 14493464 14496369 (+) 2,328 776 42,123.14 4.84 AT4G13340 CsLRX2 Csa2G004760.1 814446 817377 (+) 2,298 766 82,435.4 6.5 AT4G13340 CsLRX3 Csa3G146350.1 9732797 9733957 (+) 1,161 387 43,772.94 5.71 AT3G22800 CsPEX1 Csa6G005160.1 473143 474726 (−) 1,584 528 42,813.05 4.68 AT2G15880 CsPEX2 Csa6G006180.1 487825 489724 (−) 1,854 618 76,300.69 6.17 AT2G15880 CsPEX3 Csa6G006190.1 491897 494318 (+) 2,286 762 41,120.04 5.01 AT2G15880 BhiLRX1 Bhi01M000558 14563184 14564624 (+) 1,158 386 89,384.76 6.46 AT3G22800 BhiLRX2 Bhi02M000924 25237963 25240855 (−) 2,304 768 82,573.58 5.98 AT3G24480 BhiLRX3 Bhi06M001733 55954895 55964421 (+) 1,170 390 76,954.31 5.4 AT4G18670 BhiPEX1 Bhi09M001722 56817604 56820319 (−) 1,176 392 57,276.43 5.83 AT3G19020 BhiPEX2 Bhi12M001681 60171798 60173997 (+) 2,100 700 120,050.2 7.96 AT3G19020 ClaLRX1 Cla97C05G086020.1 4563471 4564598 (+) 1,128 376 41,548.56 4.58 AT3G22800 ClaLRX2 Cla97C06G119550.1 20517876 20520416 (+) 2,541 847 148,150.13 5.41 AT3G24480 ClaLRX3 Cla97C11G208980.1 2574856 2577150 (+) 2,295 765 166,407.81 3.7 AT4G13340 ClaPEX1 Cla97C06G112900.1 3979734 3981893 (−) 2,160 720 81,584.74 6.46 AT3G19020 CmaPEX1 CmaCh01G018740.1 12291107 12293909 (−) 1,590 530 54,054.38 5.13 AT4G33970 CmaLRX1 CmaCh05G013340.1 10085979 10089368 (−) 3,390 1,130 74,989.78 4.43 AT3G24480 CmaLRX2 CmaCh06G013780.1 8987856 8988998 (+) 1,143 381 42,903.13 5.18 AT3G22800 CmaPEX2 CmaCh08G008980.1 5563777 5568808 (−) 4,275 1,425 40,235.09 4.75 AT4G33970 CmaPEX3 CmaCh09G001990.1 826607 831562 (+) 4,956 1,652 35,186.52 4.96 AT3G19020 CmaLRX3 CmaCh10G006020.1 2780772 2788301 (−) 2,328 776 52,221.21 5.8 AT4G13340 CmaLRX4 CmaCh14G017400.1 12740774 12742300 (−) 1,527 509 88,647.32 6.16 AT3G22800 CmaPEX4 CmaCh17G004460.1 2660615 2662696 (+) 2,082 694 83,972.54 5.88 AT2G15880 CmoPEX1 CmoCh01G019320.1 13801421 13802672 (−) 1,182 394 40,916.77 4.78 AT3G19020 CmoLRX1 CmoCh06G013900.1 10200801 10201904 (+) 1,104 368 85,186.25 4.81 AT3G22800 CmoPEX2 CmoCh08G008690.1 5609995 5611160 (−) 936 312 82,799.35 6.04 AT3G19020 CmoPEX3 CmoCh08G008700.1 5621196 5622985 (−) 1,413 471 82,892.28 6.06 AT3G19020 CmoLRX2 CmoCh10G006420.1 2945225 2947750 (−) 2,526 842 42,186.25 4.73 AT4G18670 CmoLRX3 CmoCh12G012710.1 11268328 11273318 (−) 2,328 776 55,986.22 4.62 AT4G13340 CmoLRX4 CmoCh14G017780.1 13715371 13717483 (−) 1,128 376 66,641.59 5.19 AT3G22800 CmoPEX4 CmoCh17G004250.1 2835184 2837965 (+) 2,355 785 79,908.67 4.65 AT2G15880 LsiPEX1 Lsi04G016020.1 23544642 23546786 (−) 1,944 648 67,872.59 5.61 AT2G15880 LsiLRX1 Lsi05G016310.1 23953552 23954682 (−) 1,131 377 41,040.03 4.92 AT3G22800 LsiLRX2 Lsi06G008900.1 18515443 18517854 (+) 2,412 804 85,115.97 6.11 AT4G13340 LsiPEX2 Lsi09G015830.1 23966231 23967994 (−) 1,764 588 62,720.9 4.87 AT2G15880 LsiLRX3 Lsi11G002700.1 2740543 2743776 (+) 2,226 742 79,910.5 5.85 AT4G13340 MELOLRX1 MELO3C004550.2.1 28466831 28470698 (−) 1,770 590 null null AT3G24480 MELOLRX2 MELO3C006506.2.1 3799835 3800974 (+) 1,122 374 40,873.81 4.84 AT3G22800 MELOPEX1 MELO3C021195.2.1 31428500 31430511 (−) 1,947 649 70,632.34 6.37 AT3G19020 MELOPEX2 MELO3C034935.2.1 31442588 31444077 (+) 1,446 482 52,356.41 5.13 AT3G19020 CDS: coding sequence, AA: the number of amino acids, MW: Molecular weight, PI: Theoretical isoelectric point. Table 1.
Genome-wide identification of LRX gene family members in seven cucurbit species.
Figures
(7)
Tables
(1)