-
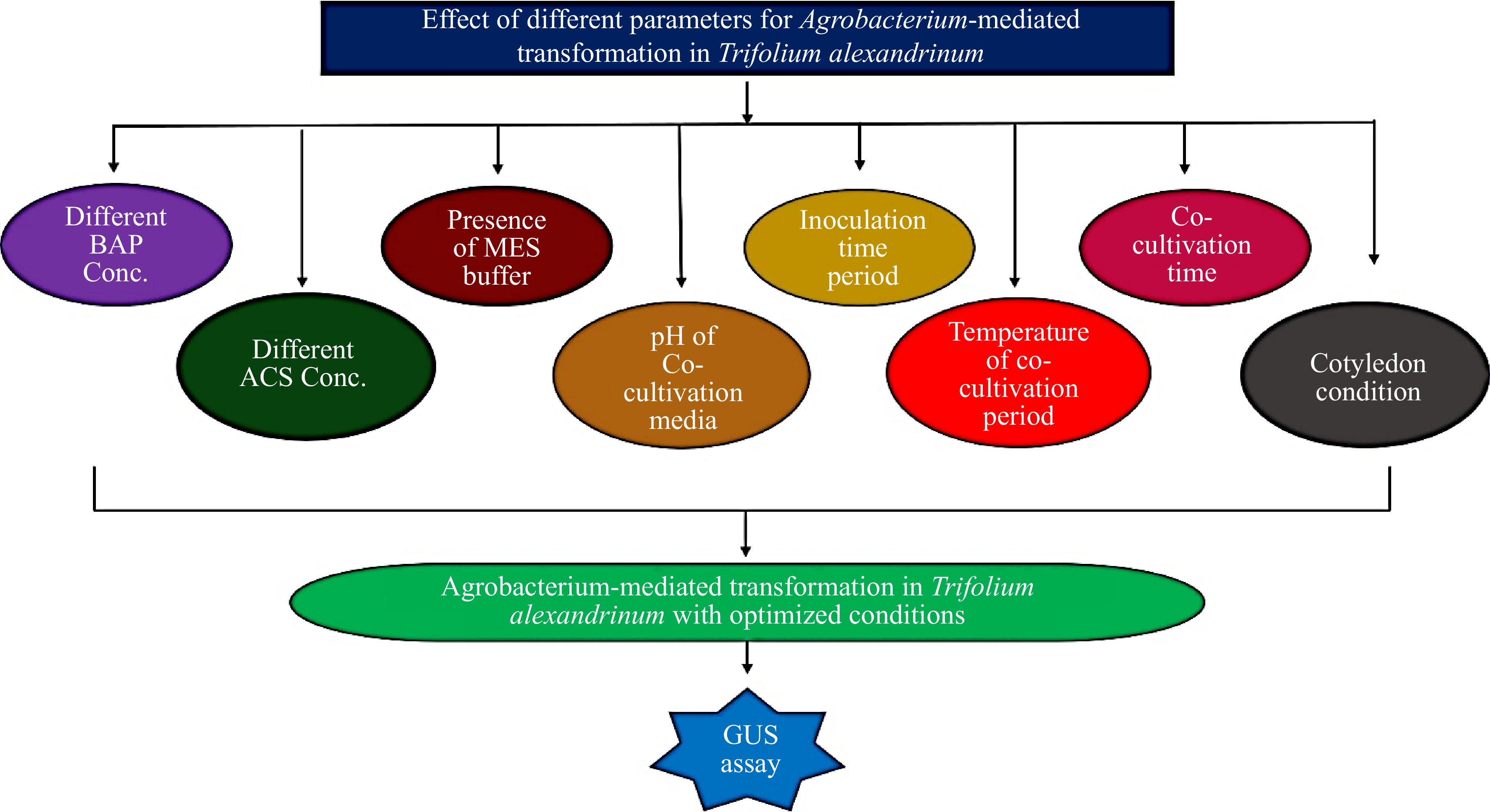
Figure 1.
Optimization of different parameters for Agrobacterium-mediated transformation of Trifolium alexandrinum.
-
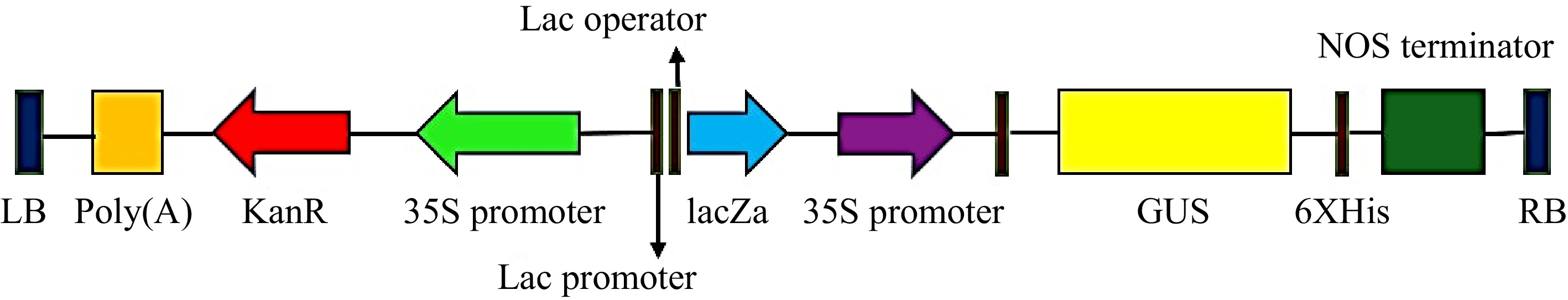
Figure 2.
T-DNA region of the plant binary vector pCAMBIA2301 used for transformation. LB: Left border, GUS: uid A, nptII: neomycin phosphotransferase-II selectable marker genes and their respective promoters plus restriction enzyme sites, NOS terminator: nopaline synthase terminator, RB: Right border.
-
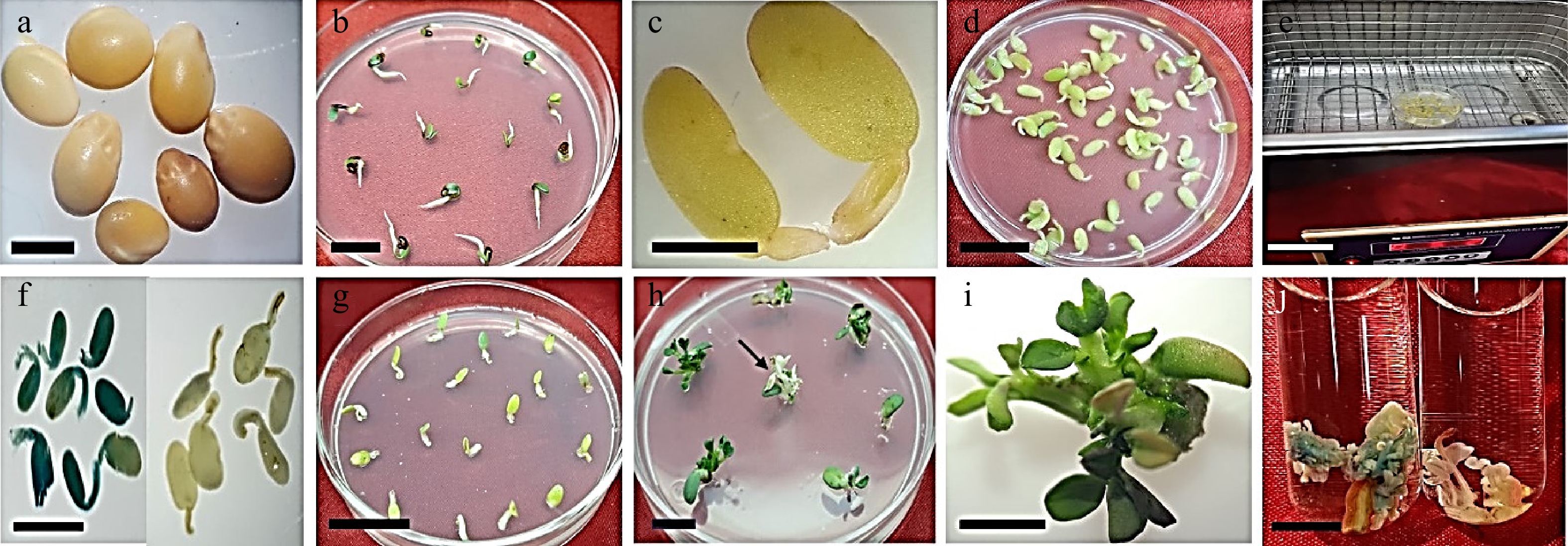
Figure 3.
A. tumefaciens mediated transformation of T. alexandrinum. (a) T. alexandrinum seeds (bar = 1 mm). (b) T. alexandrinum 3-d old seedling (bar = 10 mm). (c) Cotyledons with petiole used as explant (bar = 2 mm). (d) Explants in co-cultivation medium (bar = 10 mm). (e) Sonication of explants (bar = 6 cm). (f) Transformed explant showing transient GUS activity while untransformed (control) explant showed no GUS activity after dipping in GUS-solution, (bar = 6 mm). (g) Agrobacterium treated explants on selection media (bar = 10 mm). (h) Explants regenerating on selection medium (bar = 10 mm) and arrow showed completely bleached non-transformed shoots on selection medium. (i) Explant with multiple shoots (bar = 0.5 mm). (j) Transformed regenerated shoots showing GUS activity and non-transformed regenerated shoots without GUS activity (bar = 1 cm).
-
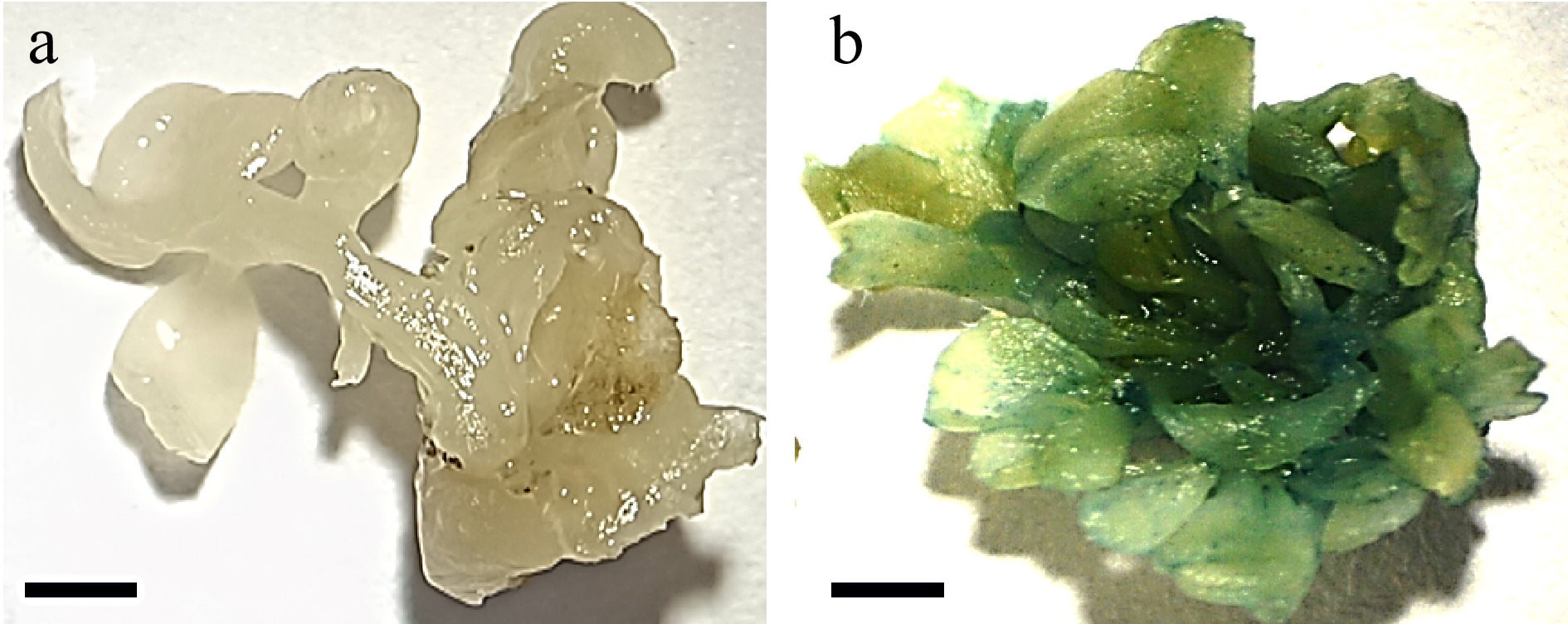
Figure 4.
Stable GUS expression in the cotyledonary explant. (a) Shoots regenerated from untransformed explant (bar = 2 mm). (b) Shoots regenerated from transformed explants showing GUS activity (bar = 2 mm).
-
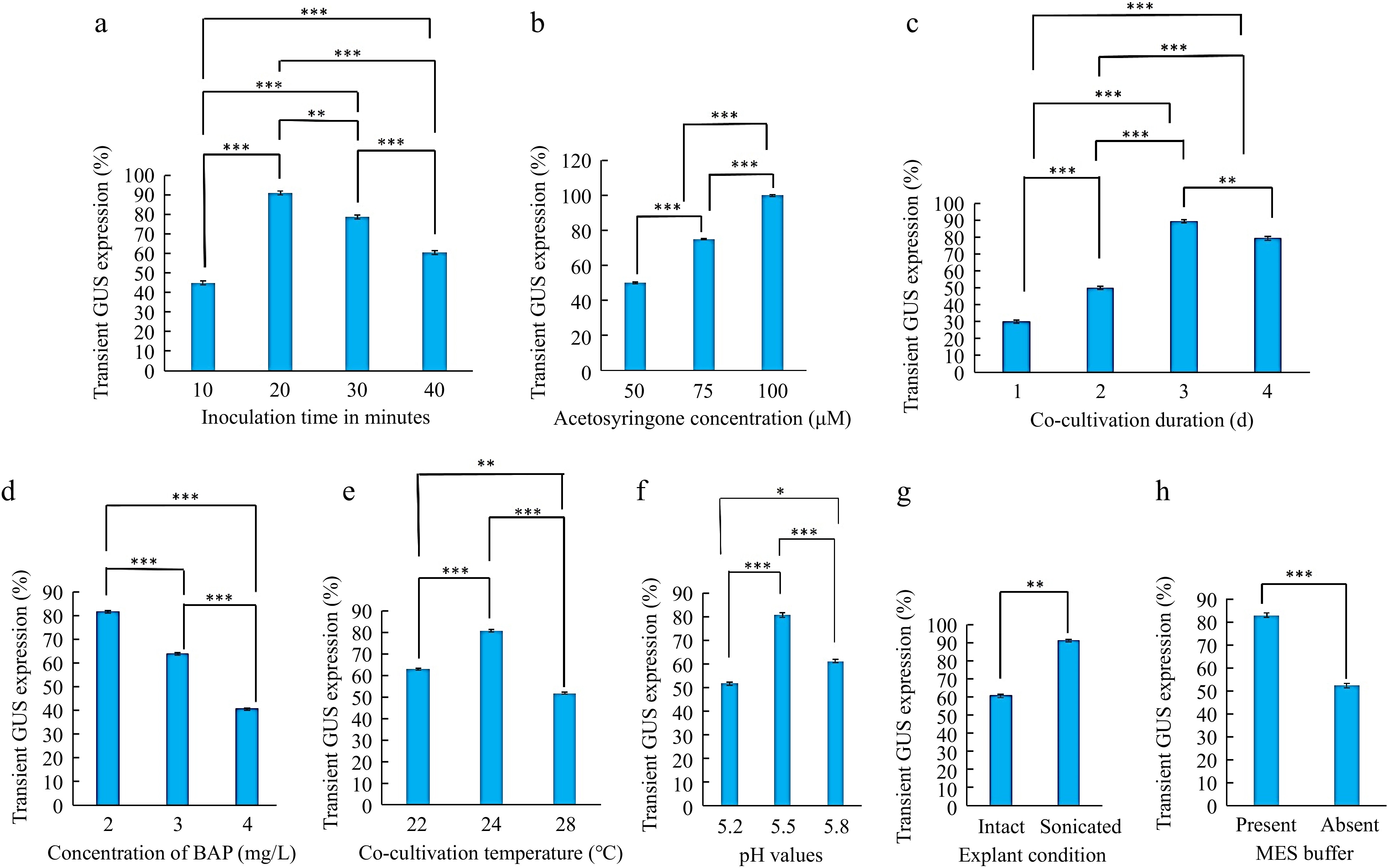
Figure 5.
Optimization of Agrobacterium-mediated genetic transformation of Egyptian clover (Trifolium alexandrinum). (a) Effect of Agrobacterium inoculation time in minutes. (b) Effect of different acetosyringone concentrations (μM). (c) Effect of co-cultivation duration. (d) Effect of BAP concentrations in cocultivation medium. (e) Effect of temperature during co-cultivation. (f) Effect of co-cultivation medium pH. (g) Effect of explant condition (intact and sonicated). (h) Effect of presence of MES buffer in cocultivation medium. Significant differences were obtained at * p < 0.05, ** p < 0.01, *** p < 0.001 levels.
-
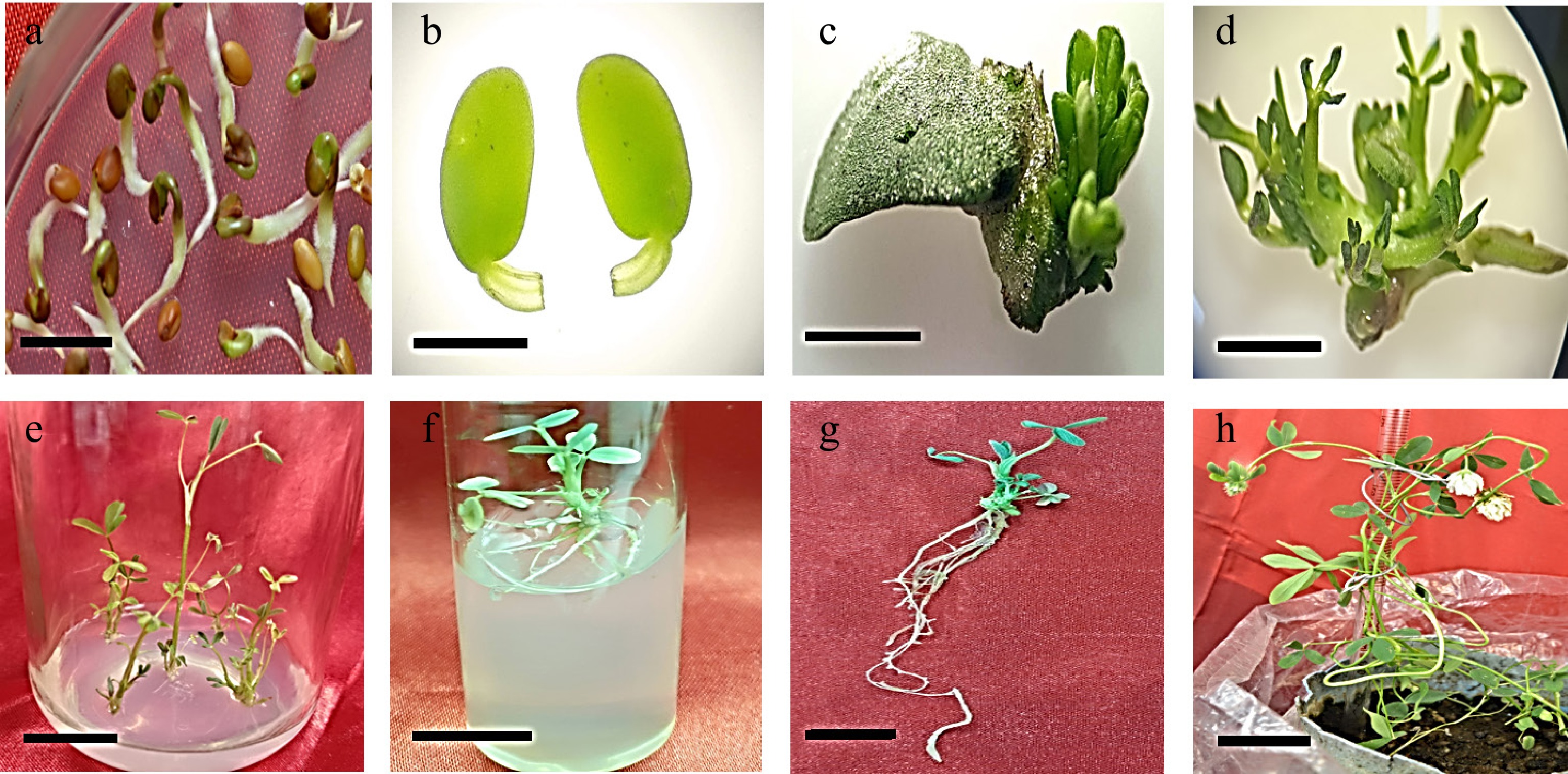
Figure 6.
Development of Trifolium alexandrinum plants. (a) Three-day-old seedling, (bar = 10 mm), (b) excised cotyledonary explant with petiole (bar = 2 mm), (c), (d) young multiple shoots emerging from the petiolar region of cotyledon explants (bar = 0.3 cm, bar = 0.7 cm), (e) multiple shoots arising from cotyledon explant (bar = 0.8 cm), (f) elongated shoots on elongation medium (bar = 2 cm), (g) rooted shoot on rooting medium (bar = 2 cm), (h) mature plant with white flowers (bar = 9 cm).
-
S. no. Species Explant Gene transferred Vector and Agrobacterium strain Transformation efficiency Selection Analysis Ref. 1 Trifolium
AlexandrinumCotyledon uid A gene p7i-UG (EHA105) − − GUS assay [1] 2 Trifolium
repens− WXP1 and GUS pCAMBIA3301
(AGL1)− Phosphinothricin GUS staining, Northern blot, PCR and RT-PCR [8] 3 Trifolium occidentale Cotyledon Bar selection gene and uid A gene pHZBar-intGUS (GV3101) 7.5% Ammonium glufosinate and Timentin Southern blot and GUS assay [9] 4 Trifolium subterranean Hypocotyl Bar, uid A, nptII, $ \propto -al $ pMCP3
(AGL1)− Phosphinothrycin Southern blotting, Nothern blotting, GUS assay, and α-AI immunoblot assay [10] 5 Trifolium subterranean Cotyledon uid A gene and hyg gene pH35
(AGL0)5.2% Hygromycin and Cefotaxime PCR [11] 6 Trifolium
PratenseCallus generated from cotyledon and hypocotyl IFS gene pRI101-AN-IFS
(LBA4404)− Kanamycin and cephalosporin RT-PCR [12] Table 1.
Trifolium species with explant used, gene transferred, antibiotic for selection, the vector used, analysis method, and transformation efficiency.
Figures
(6)
Tables
(1)