-
Trifolium alexandrinum (Berseem) earns its moniker as the Fodder Crop Champion due to its exceptional nutrient profile, wide adaptability, and the ability to be harvested multiple times[1,2]. Typically sown in October, it yields between 100 and 150 tons of fresh biomass per hectare over five to six harvests extending through November and May. Its remarkable nitrogen-fixing capability significantly enhances soil fertility by fixing 297−400 kg of atmospheric nitrogen per hectare[1]. Trifolium incorporation into crop rotations plays a pivotal role in enriching the soil with nitrogen to prepare it for subsequent summer crops like cotton and rice, necessitating planting at least once every two years[3]. It is recognized as a premier Rabi (winter season) forage crop in the entire North West Zone, Hill Zone, and portions of the Central and Eastern Zones of India, it spans over 20 lakh hectares[4]. Efficient in vitro plant regeneration and transformation systems are imperative for successfully introducing desired foreign genes into plants. Low genetic variations pose a significant obstacle to the advancement of forage legumes, necessitating genetic transformation to introduce new genes or cultivate new varieties. Biotechnological approaches offer potent alternatives for enhancing crop genetics, including elevating nutritional content and bolstering resistance to biotic and abiotic stresses. While progress has been made in creating genetic variations in T. alexandrinum, genetic transformation remains limited in this legume fodder crop, especially for Indian varieties due to genotype-dependent low regeneration frequency, poor susceptibility to Agrobacterium, and response to selective agents[1,3,5]. Only a few reports are available on this species in vitro culture and genetic transformation[6,7] (Table 1). The proper selection of suitable explants, optimization of Agrobacterium infection and regeneration conditions can overcome the limitations in the genetic transformation of this species. Therefore, in this study, the various parameters affecting genetic transformation were optimized using transient GUS activity in cotyledon explants (Fig. 1) to establish a reliable, and efficient Agrobacterium-mediated genetic transformation of T. alexandrinum to improve its high biomass production, resilience to adverse climatic and biotic stresses, substantial nitrogen fixation and more nutritious and palatable fodder.
Table 1. Trifolium species with explant used, gene transferred, antibiotic for selection, the vector used, analysis method, and transformation efficiency.
S. no. Species Explant Gene transferred Vector and Agrobacterium strain Transformation efficiency Selection Analysis Ref. 1 Trifolium
AlexandrinumCotyledon uid A gene p7i-UG (EHA105) − − GUS assay [1] 2 Trifolium
repens− WXP1 and GUS pCAMBIA3301
(AGL1)− Phosphinothricin GUS staining, Northern blot, PCR and RT-PCR [8] 3 Trifolium occidentale Cotyledon Bar selection gene and uid A gene pHZBar-intGUS (GV3101) 7.5% Ammonium glufosinate and Timentin Southern blot and GUS assay [9] 4 Trifolium subterranean Hypocotyl Bar, uid A, nptII, $ \propto -al $ pMCP3
(AGL1)− Phosphinothrycin Southern blotting, Nothern blotting, GUS assay, and α-AI immunoblot assay [10] 5 Trifolium subterranean Cotyledon uid A gene and hyg gene pH35
(AGL0)5.2% Hygromycin and Cefotaxime PCR [11] 6 Trifolium
PratenseCallus generated from cotyledon and hypocotyl IFS gene pRI101-AN-IFS
(LBA4404)− Kanamycin and cephalosporin RT-PCR [12] -
Mature seeds of T. alexandrinum variety Mescavi were obtained from the Chaudhary Charan Singh Haryana Agriculture University, Hisar, Haryana, India. Agrobacterium tumefaciens strain EHA105 harboring a binary vector pCAMBIA2301 that carries a scorable marker, uid A gene encoding for β glucuronidase (GUS), and a selectable marker, nptII gene encoding for neomycin phosphotransferase was used for transformation. Both genes are controlled by CaMV35S promoter (Fig. 2).
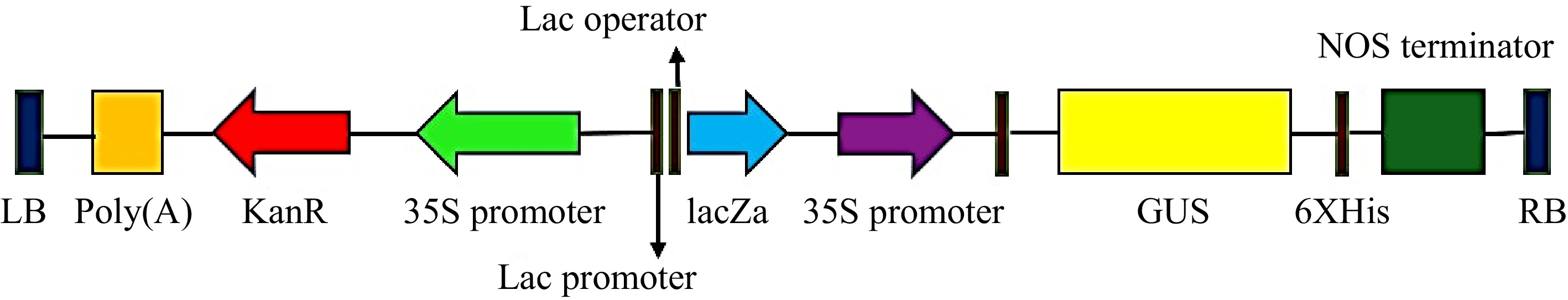
Figure 2.
T-DNA region of the plant binary vector pCAMBIA2301 used for transformation. LB: Left border, GUS: uid A, nptII: neomycin phosphotransferase-II selectable marker genes and their respective promoters plus restriction enzyme sites, NOS terminator: nopaline synthase terminator, RB: Right border.
Seed germination and preparation of explants
-
Healthy seeds were surface sterilized by stirring them in an aqueous 0.2% mercuric chloride solution for 3 min in a laminar air-flow chamber. Afterward, the seeds were washed five to six times with autoclaved distilled water to remove all traces of mercuric chloride. The seeds were then germinated on MSB5 medium[13] containing salts of Murashige and Skoog medium and vitamins of B5 medium[14], and 3% sucrose for 3 d at 24 ± 2 °C under a 16-h photoperiod of cool-white fluorescent light with an intensity of 80 μEm−2·s−1.
Preparation of explants, culture medium, and culture conditions
-
The cotyledon explants with petioles (~6 mm in length) were cut closely to the embryonic axis using a sterilized scalpel and forceps inside a laminar airflow cabinet. The cotyledons with intact petioles were cultured on MSB5 semi-solid media with varied amounts of BAP (2−4 mg/L) by slightly embedding the proximal cut end in the medium. The pH of the medium was maintained at 5.8 before autoclaving for 20 min at 121 °C and 15 psi. The cultures were kept at 24 ± 2 °C and exposed to 16 h of cool-white fluorescent light.
Optimization of parameters affecting transformation
-
Several variables affecting transformation efficiency were optimized using transient GUS activity to establish an efficient transformation protocol for T. alexandrinum. Three-day-old cotyledon explants excised from in vitro-raised seedlings were incubated in A. tumefaciens (pCAMBIA2301) solution and/or sonicated in a bath-type sonicator (at 40 kHz) for 30 s and gently rotated at 80 rpm for 10–40 min (Fig. 3). The explants were air-dried on sterile filter paper and co-cultivated on filter paper hydrated with liquid MSB5 co-culture medium containing MES buffer, 50−100 μM acetosyringone, 0−4 mg/L BAP at 5.2−5.8 pH under 22−28 °C and dark conditions for 1−4 d. Sixty explants were used for each experimental parameter and each experiment was carried out in triplicate (Fig. 4).
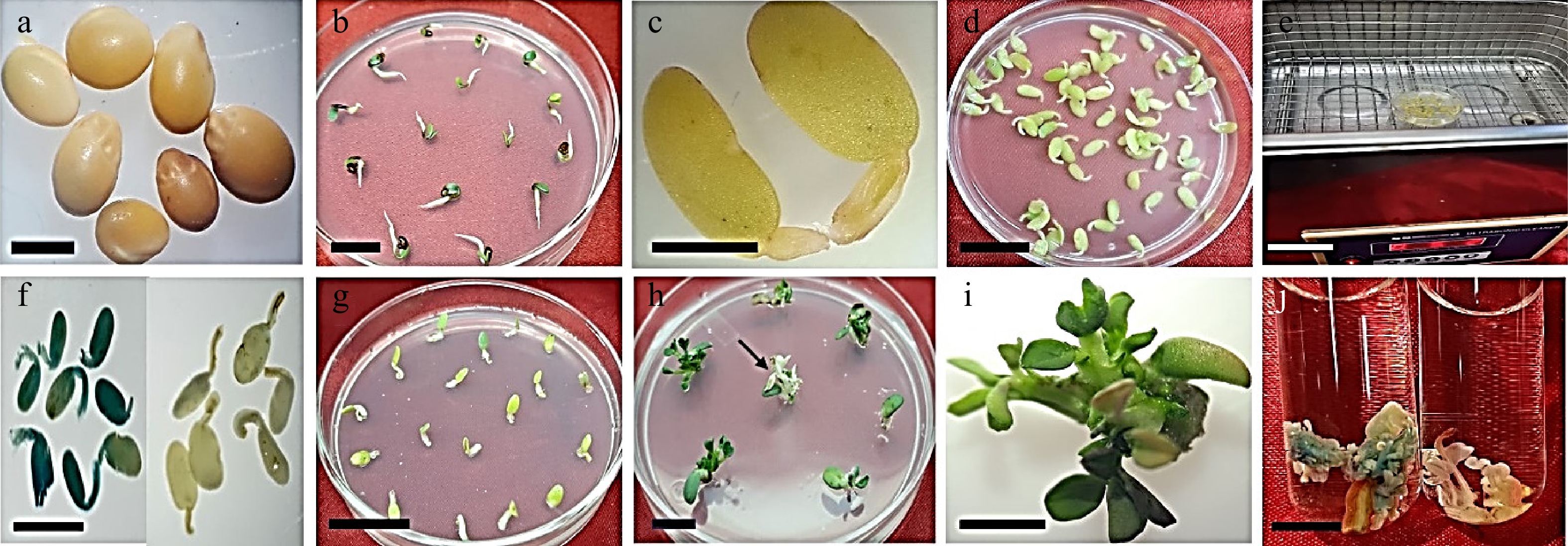
Figure 3.
A. tumefaciens mediated transformation of T. alexandrinum. (a) T. alexandrinum seeds (bar = 1 mm). (b) T. alexandrinum 3-d old seedling (bar = 10 mm). (c) Cotyledons with petiole used as explant (bar = 2 mm). (d) Explants in co-cultivation medium (bar = 10 mm). (e) Sonication of explants (bar = 6 cm). (f) Transformed explant showing transient GUS activity while untransformed (control) explant showed no GUS activity after dipping in GUS-solution, (bar = 6 mm). (g) Agrobacterium treated explants on selection media (bar = 10 mm). (h) Explants regenerating on selection medium (bar = 10 mm) and arrow showed completely bleached non-transformed shoots on selection medium. (i) Explant with multiple shoots (bar = 0.5 mm). (j) Transformed regenerated shoots showing GUS activity and non-transformed regenerated shoots without GUS activity (bar = 1 cm).
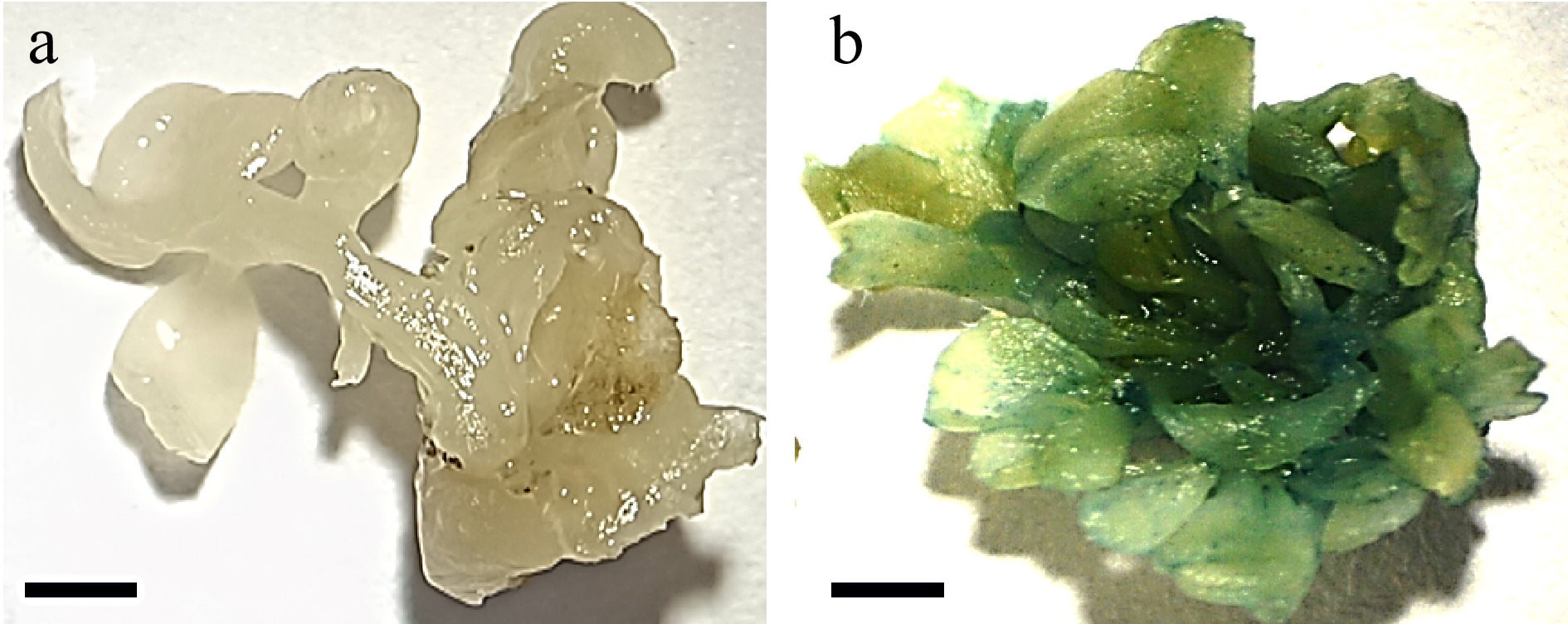
Figure 4.
Stable GUS expression in the cotyledonary explant. (a) Shoots regenerated from untransformed explant (bar = 2 mm). (b) Shoots regenerated from transformed explants showing GUS activity (bar = 2 mm).
Transient and stable GUS gene assay
-
The cocultured explants were thoroughly washed with sterilized distilled water and a final wash with cefotaxime antibiotic to kill all bacteria attached to the surface of the explants, and dried on a sterile filter paper. The explants were either incubated in a freshly prepared X-Gluc solution at 37 °C for approximately 24 h[15] or transferred onto MSB5 medium containing 2 mg/L BAP for shoot regeneration under culture conditions as mentioned in the seed germination section. The explants were treated with 70%−90% ethanol to remove their chlorophyll and examined under a stereo-zoom microscope. The frequency of GUS activity was determined by dividing the number of cotyledon explants showing blue color with the total number of cotyledon explants incubated in X-Gluc.
Statistical data analysis
-
Statistical analysis was performed for each parameter and experiment using one-way ANOVA in Graph Pad Prism 9.4. Data was recorded on 60 explants per analysis and was replicated three times. The 'Honest Significant Difference' test by Tukey was utilized to determine major variations among different groups at various levels of significance, * p < 0.05, ** p < 0.01, *** p < 0.001.
Effect of inoculation time with bacterial suspension
-
The cotyledons with petiole explants were cut from young seedlings and incubated with the co-culture medium. The number of explants that expressed GUS following Agrobacterium infection depends on the co-incubation time. In this period, Agrobacterium had the opportunity to invade meristematic cells. This study examined various (20, 30, and 40 min) co-cultivation periods.
Effect of acetosyringone concentration
-
Naturally, wounded plant cells release a phenolic compound, acetosyringone, which stimulates Agrobacterium for attachment with wounded plant cells and induces the Vir gene. Vir gene expression controls the T-DNA transfer in the plant cell[16]. It has been demonstrated that GUS is more frequently used for the genetic transformation of many plant species. The effect of different acetosyringone concentrations (50, 75, and 100 μM) on transient GUS expression was investigated by co-cultivating cotyledonary explant for 3 d in the dark.
Effect of the co-cultivation period
-
The co-cultivation incubation period influenced the genetic transformation. In legume (Cicer arietinum), the co-cultivation period was carried out for 1–4 d and optimized that 2 d of co-cultivation was better for transformation[17]. A short period is not beneficial because bacteria need sufficient time to stick to and invade plant cells. In contrast, a long period causes necrosis and, as a result, reduces T-DNA transfer. In this study, explants were incubated in co-cultivation media (pH 5.5) in the dark for 2−4 d to optimize the co-cultivation period for transient genetic expression in Trifolium alexandrinum.
Effect of BAP concentration
-
In earlier studies[18,19], it was underlined how crucial it was to include phytohormone in the co-culture medium to increase transformation incidence. A thorough investigation found that phytohormone in the medium may aid the transformants to survive in intense stress brought on by wounding and exposure to Agrobacterium. The most common compound utilized in different legume species was 6-benzyl aminopurine (BAP)[20]. For maximum transformation efficiency in Trifolium alexandrinum, different concentrations of BAP (2, 3, and 4 mg/L) were used with MsB5 media.
Effect of co-cultivation temperature
-
The Agrobacterium-mediated transformation was affected by co-cultivation temperature in Trifolium pratense[12]. Lower temperature enhanced pilus construction and increased the number of pili on the cell surface[21]. The enhanced transformation may have been partly caused by the T-DNA transfer machinery's Vir B-Vir D4 components and improved performance at low temperatures[22]. In the present study, the co-cultivation temperature was optimized between 22−24 °C to increase the transformation efficacy.
Effect of co-cultivation medium pH
-
The pH level of the co-cultivation medium plays a pivotal role in determining the virulence and transformation efficiency of Agrobacterium. The expression of Vir genes is influenced by the pH of the surrounding environment, with optimal transformation efficiency observed within the pH range of 5.0 to 6.0 in the co-cultivation medium[23]. To investigate this, three specific pH values (5.2, 5.5, and 5.8) were assessed, each with approximately 60 explants assigned to the respective treatment groups.
Effect of cotyledon explant condition
-
For genetic transformation in Trifolium, cotyledonary explants having meristematic cells are the primary goal, and mechanical damage to the meristematic region increases the potential of foreign gene uptake. Making tiny wounds and sonication enables Agrobacterium to enter meristematic cells quickly. The transformation effectiveness gradually increased with sonication cycles lasting up to 30 s.
Effect of MES buffer
-
MES buffer initiates the Agrobacterium-mediated genetic transformation[24]. In the present study, the impact of MES buffer, either being present or not in the transient GUS experiment was investigated.
GUS gene assay
-
The explants were co-cultured in liquid MSB5 co-culture medium for 1−3 d at 22−28 °C under dark conditions, were washed with sterilized distilled water and antibiotic to clean extra bacteria attached on the surface of explants, and later patted dry on a sterile filter paper, and incubated in X-Gluc dye at 37 °C for approximately 24 h[15]. The tissue should be de-coloured in 70%−90% ethanol the day after the assay solution is removed. There were 60 explants employed for each experimental parameter, and each experiment was carried out in triplicate.
$\begin{split}& \rm Frequency\; of\; GUS\; expression = \\&\rm\dfrac{The \;number \;of \;cotyledon\; explants\; showing\; a\; blue\; color}{Total\; number\; of\; cotyledon \;explants\; incubated \;in \;X{\text-}Gluc}\end{split}$ Where the gene has been actively expressed can be seen as remarkable blue spots by the GUS-induced stain. Thus, robust promoter activity results in significant staining, whereas weak promoter activity results in minimal staining. Optimized Agrobacterium transformation protocol led to a high frequency of transformation (up to 96%) and yielded high blue spots on explants after following selected best parameters that effect transformation. Microscopic observation of the blue spots revealed transient uid A gene incorporation into the explant; besides, no blue spots are visible in control explants (without Agrobacterium treatment) (Fig. 4). Thus, following these optimized parameters gave the highest transformation efficiency in Trifolium alexandrinum.
-
Agrobacterium-mediated genetic transformation has been the most common method for Trifolium species transformation[9−11,25]. A. tumefaciens and A. rhizogenes have been used for genetic transformation[5,26]. However, an efficient Agrobacterium-mediated genetic transformation for an Indian cultivar of T. alexandrinum has very few reports. The barriers to efficient gene transfer in this legume can be overcome with methodological adjustments, particularly in co-cultivation conditions and explant selection. The significant improvement in the efficiency of Agrobacterium-mediated transformation of T. alexandrinum presents a promising avenue for genetic improvement of this species.
Effect of inoculation time with bacterial suspension
-
Three-day-old cotyledon explants with petiole incubated in Agrobacterium suspension for 10, 20, 30, and 40 min showed significant differences in transient GUS expression. Explants incubated for 20 min in Agrobacterium suspension exhibited a maximum frequency of GUS expression, i.e. 91.11%, and those with higher incubation time, i. e., 30- and 40-min reduced percent explants with GUS expression. Therefore, the incubation period of 20 min was optimal in cotyledonary explants of T. alexandrinum (Fig. 5a), and a similar effect was observed in Trifolium repens[27], Vigna mungo L. Hepper[28]. Further increase in incubation time, decreased the transformation frequency with excessive bacteria growth that were difficult to eliminate. In some other plant species and legumes, explants were incubated for 20−40 min[16,27,29].
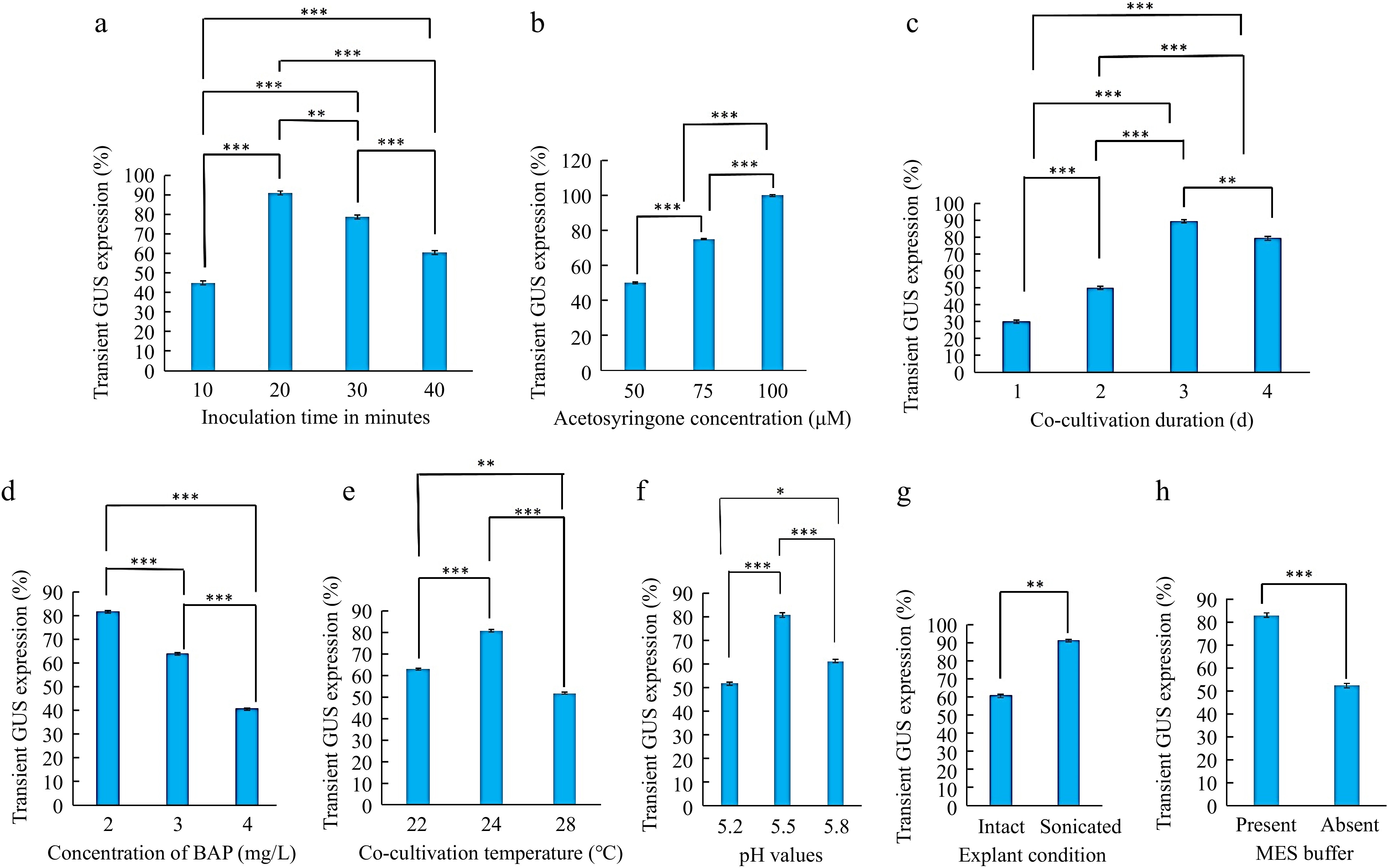
Figure 5.
Optimization of Agrobacterium-mediated genetic transformation of Egyptian clover (Trifolium alexandrinum). (a) Effect of Agrobacterium inoculation time in minutes. (b) Effect of different acetosyringone concentrations (μM). (c) Effect of co-cultivation duration. (d) Effect of BAP concentrations in cocultivation medium. (e) Effect of temperature during co-cultivation. (f) Effect of co-cultivation medium pH. (g) Effect of explant condition (intact and sonicated). (h) Effect of presence of MES buffer in cocultivation medium. Significant differences were obtained at * p < 0.05, ** p < 0.01, *** p < 0.001 levels.
Effect of acetosyringone concentration
-
The addition of a phenolic compound, acetosyringone (50, 75, and 100 μM) in the cocultivation medium significantly increased the transformation efficacy with an increase in its concentration. The maximum GUS expression frequency (90%) was observed at 100 μM of acetosyringone (Fig. 5b). The presence of acetosyringone in the bacterial suspension and co-cultivation medium is essential for the T. alexandrinum notable transformation. Researchers also successfully used acetosyringone at a concentration of 100 μM to increase transformation efficiency like in Cicer arietinum, Citrullus lematus Pisum sativum, and Vigna unguiculata L. Walp[16,29−31] whereas in T. subterranean and Sesamum indicum 20 μM acetosyringone was used for genetic transformation[10,32].
Effect of co-cultivation period
-
The Agrobacterium-inoculated explants were co-cultured for 1−4 d in the dark. The co-cultivation duration had a significant effect on GUS expression frequency. The 3-d co-cultivation period was the most effective, with a high transformation frequency (89.4%) for T. alexandrinum (Fig. 5c) as the same co-cultivation period was used in Vigna unguiculata L. Walp, Vigna radiata[31,33]. Co-cultivation for short periods of 1 and 2 d was not sufficient to improve GUS gene expression and extending the cocultivation beyond 4 d caused necrosis due to the excessive growth of Agrobacterium resulting in a decline of transformation frequency and explants regeneration. Most Trifolium species like T. subterranean[10], T. pratense[12], and other legumes[34,35] required a 2−7-d co-cultivation period for the maximum transformation frequency.
Effect of BAP concentration
-
As reported earlier, BAP was the most effective cytokinin for shoot regeneration in Trifolium species[1,3]. In this study, explants incubated with different BAP concentrations, 2, 3 and 4 mg/L showed significant differences in transient GUS expression. BAP might have helped the explants to survive under bacterial stress conditions. Maximum transformation efficacy (81.66%) was observed in those explants incubated on a co-cultivation medium with 2 mg/L BAP (Fig. 5d). TDZ, kinetin, and zeatin were also used in different concentrations in some other plant species[10,12,36].
Effect of co-cultivation temperature
-
Co-cultivation at 25 °C gave the maximum transitory GUS expression in Trifolium subterraneum[10] and at 26 °C produced the best transformation in both Trifolium species, T. repens, T. pratense, and Medicago[37]. Among the three temperatures (22, 24, and 28 °C) evaluated in the current study, T. alexandrinum cotyledonary explants transformation had the maximum effectiveness, i.e., 80.55% GUS expression at 24 °C, (Fig. 5e) indicating that a suitable low temperature during co-cultivation can enhance Agrobacterium-mediated transformation.
Effect of co-cultivation medium pH
-
In the present study, the effect of the co-cultivation medium at different pH (5.2, 5.5, and 5.8) was tested on transient GUS activity. The maximum GUS transformation frequency of 80.55% was at pH 5.5 (Fig. 5f) while the minimum frequency of 51.6% was at pH 5.2. Co-cultivation medium pH is the essential factor responsible for the activation of vir gene expression. So, the pH should be adjusted appropriately when preparing a co-cultivation medium. pH 5.3 and 5.4, were also used in genetic transformation studies in Trifolium alexandrinum and Cowpea[7,31].
Effect of sonication of cotyledon explants
-
Sonication of the cotyledonary explants at 40 kHz for 30 s significantly improved the transformation efficiencies to 91.1% which was 1.5-fold of the non-sonicated (intact) explants (Fig. 5g). However, sonication treatment longer than 30 s reduced the transformation efficiency. Sonication treatments considerably boosted the efficacy of genetic transformation in several plant species[38]. More phenolic compounds may be released from the micro-wounds made by sonication, enabling Agrobacterium to enter the tissue deeper and increase the effectiveness of plant transformation[39,40].
Effect of MES buffer
-
The pH of the Agrobacterium inoculation and co-cultivation medium affected the transformation efficacy of T. alexandrinum in the present study. The use of MES buffer in the co-cultivation medium maintained or retained the pH and without MES buffer use, the pH was changed to the lower side[41]. MES buffer improves the attachment of Agrobacterium to the plant leading to better T-DNA transfer and integration[16]. The MES buffer presence in the cocultivation medium improved the transformation efficiency by 82.7% (Fig. 5h) than without an MES buffer, it was only 52.2%. In the genetic transformation of several legumes, an MES buffer is also added to the cocultivation medium.
Development of healthy T. alexandrinum plant
-
Cocultured 3-day-old cotyledonary explants with petiole (Fig. 6a, b) on transfer onto shoot regeneration medium (MS medium having 2 mg/L BAP) containing 80 mg/L kanamycin and 250 mg/L cefotaxime regenerated healthy shoots (Fig. 6c, d). The regenerated shoots were elongated on MS medium containing 0.15 mg/L BAP, 80 mg/L kanamycin, and 250 mg/L cefotaxime (Fig. 6e). The elongated shoots were transferred to MS medium containing 1.5 mg/L IBA, 200 mg/L cefotaxime, and 30 mg/L kanamycin for rooting (Fig. 6f, g). Rooted shoots were grown in pots containing sterilized soil for acclimatization and flowering (Fig. 6h).
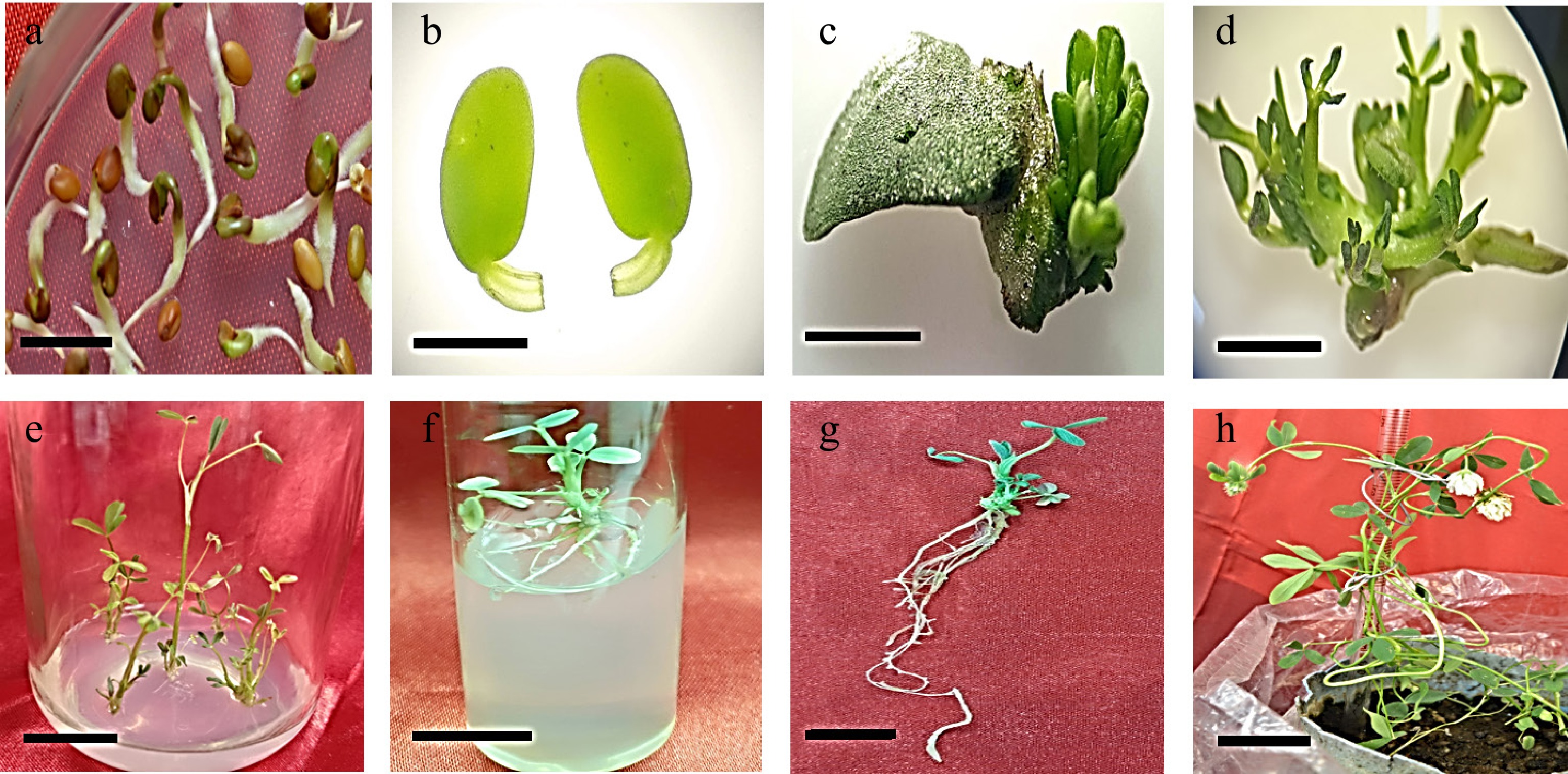
Figure 6.
Development of Trifolium alexandrinum plants. (a) Three-day-old seedling, (bar = 10 mm), (b) excised cotyledonary explant with petiole (bar = 2 mm), (c), (d) young multiple shoots emerging from the petiolar region of cotyledon explants (bar = 0.3 cm, bar = 0.7 cm), (e) multiple shoots arising from cotyledon explant (bar = 0.8 cm), (f) elongated shoots on elongation medium (bar = 2 cm), (g) rooted shoot on rooting medium (bar = 2 cm), (h) mature plant with white flowers (bar = 9 cm).
-
Optimizing factors that enhance Agrobacterium virulence for T-DNA transport and improves the ability to survive and regenerate transformed cells is crucial for legume crops like Berseem. In the current study, standardized parameters have been established for an efficient Agrobacterium-mediated transformation strategy, focusing on T. alexandrinum cotyledonary explants. Through experimentation with various variables, including bacterial inoculation time, co-cultivation duration, acetosyringone concentration, sonication-induced injuries, co-cultivation medium pH, presence of MES buffer, BAP concentrations, and co-cultivation temperature, we observed increased transformation frequencies. After thorough optimization, it was determined that cotyledonary explants with petioles, subjected to injury and inoculated in a co-cultivation medium with a pH of 5.5 for 20 min, supplemented with acetosyringone (100 μM), BAP (2 mg/L), MES buffer, and incubated for 3 d at 24 ± 2 °C, exhibited enhanced transformation efficiency. The inclusion of MES buffer maintained a constant pH in the co-cultivation medium. This optimized procedure holds promise for efficient genetic modification of Trifolium alexandrinum and other leguminous plants.
Despite advancements, challenges persist in achieving optimal transformation efficiency in T. alexandrinum. This paper explores potential future directions and strategies to overcome these challenges, ultimately advancing the genetic improvement of this valuable forage crop.
Understanding host-pathogen interactions
-
Investigating the molecular mechanisms underlying the interaction between T. alexandrinum and Agrobacterium tumefaciens is crucial. A comprehensive understanding of factors influencing transformation efficiency, such as host cell receptivity, bacterial virulence, and genetic compatibility, will inform targeted optimization strategies.
Enhancing vector design
-
Development of improved binary vectors tailored specifically for T. alexandrinum transformation can enhance efficiency. This involves optimizing promoter and terminator sequences, incorporation of tissue-specific regulatory elements, and exploring alternative selectable marker genes to minimize pleiotropic effects and increase transformation frequency.
Exploring novel Agrobacterium strains
-
Screening and characterization of diverse Agrobacterium strains for their suitability in T. alexandrinum transformation could lead to the identification of strains with enhanced virulence and transformation efficiency. Additionally, engineering Agrobacterium strains to improve their capacity for DNA delivery and integration into the plant genome could be a promising avenue.
Refining tissue culture protocols
-
Fine-tuning tissue culture conditions, including explant type, hormone concentrations, and co-cultivation parameters, can significantly impact transformation efficiency. Optimization of these parameters based on the specific requirements of T. alexandrinum will contribute to consistent and reproducible transformation outcomes.
Incorporating CRISPR-Cas technology
-
Integration of CRISPR-Cas genome editing technology with Agrobacterium-mediated transformation can facilitate precise genetic modifications in T. alexandrinum. Harnessing CRISPR for targeted gene knockouts, gene editing, and regulatory element manipulation offers unprecedented opportunities for trait improvement with high specificity and efficiency.
Utilizing omics technologies
-
Leveraging transcriptomics, proteomics, and metabolomics approaches can provide insights into the molecular responses of T. alexandrinum to transformation and identify key regulatory nodes. Integrating omics data with transformation experiments enables a holistic understanding of the genetic and biochemical pathways involved, guiding rational design strategies for enhanced transformation efficiency.
Studying epigenetic regulation
-
Investigating the role of epigenetic modifications in modulating gene expression during T. alexandrinum transformation could uncover epigenetic targets for manipulation to improve transformation efficiency and stability of transgene expression.
Addressing biosafety concerns
-
As genetic modification technologies advance, addressing biosafety and regulatory considerations are paramount. Proactive engagement with regulatory bodies and stakeholders, along with thorough risk assessment of transformed T. alexandrinum lines ensures responsible deployment and acceptance of genetically modified crops.
In conclusion, advancing the efficiency of Agrobacterium-mediated transformation in T. alexandrinum requires a multidisciplinary approach integrating molecular biology, microbiology, bioinformatics, and plant physiology. By addressing key challenges and implementing innovative strategies, researchers can unlock the full potential of genetic transformation for improving this vital forage crop.
-
The authors confirm contribution to the paper as follows: performing experiment and writing manuscript: Prajapati M, Chaudhary D; assisting in experiment and manuscript: Ahlawat YK, Jaiwal PK, Jaiwal R; project supervision: Chaudhary D, Jaiwal PK; study conception: Ahlawat YK, Chaudhary D. All authors reviewed the results and approved the final version of the manuscript.
-
All data generated or analyzed during this study are included in this published article.
Mukta Prajapati and Darshna Chaudhary are grateful to the ClR-SRF and Department of Biotechnology, New Delhi for the financial support (09/382(0235)/2019-EMR-1 and BT/INF/22/SP43043/2021) respectively.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Prajapati M, Chaudhary D, Jaiwal PK, Jaiwal R, Ahlawat YK. 2024. Optimizing Agrobacterium-mediated transformation efficiency in an Indian cultivar of Trifolium alexandrinum L. Grass Research 4: e019 doi: 10.48130/grares-0024-0018
Optimizing Agrobacterium-mediated transformation efficiency in an Indian cultivar of Trifolium alexandrinum L.
- Received: 07 May 2024
- Revised: 21 August 2024
- Accepted: 27 August 2024
- Published online: 10 September 2024
Abstract: Trifolium alexandrinum, commonly known as Egyptian clover, plays a crucial role as a forage crop of significant agricultural importance. Despite its importance, achieving efficient genetic transformation in this species has remained a challenge, hindering potential advancements in its improvement. In this study, numerous parameters, e.g. the Agrobacterium-inoculation time and cocultivation duration, acetosyringone and BAP concentrations, and the presence of MES buffer in the co-cultivation medium, and its pH and temperature during co-cultivation impacting the transformation efficiency were methodically investigated using cotyledons with petiole as an explant. The cotyledon explants without mechanical injury before inoculation with Agrobacterium tumefaciens for 20 min and co-cultivation for 3 d on a medium containing MES buffer, 2 mg/L BAP, and 100 μM acetosyringone with a pH of 5.5 at 24 °C resulted in GUS activity in 60% of the explants. In contrast, those mechanically injured with sonication showed the maximum GUS activity in 91.11%. Subsequently, the co-cultivated explants were regenerated on an MS medium with 2 mg/L BAP. Eighty percent of the explants developed multiple shoots from the petiolar end of the cotyledons. These regenerating explants with multiple shoots showed stable GUS activity. The optimized transformation protocol with enhanced efficiency would broaden genetic manipulation capabilities in T. alexandrinum by gene editing technologies and synthetic biology approaches.
-
Key words:
- Trifolium alexandrinum /
- Agrobacterium /
- GUS /
- Genetic transformation /
- Acetosyringone