-
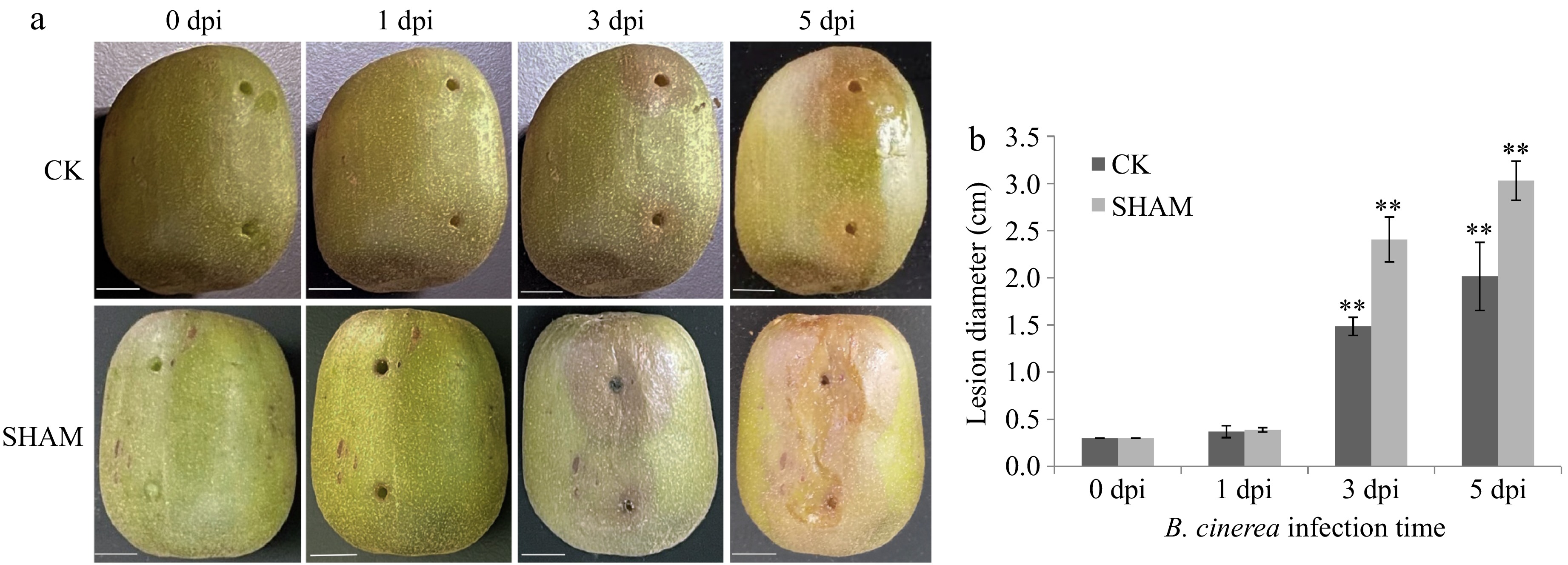
Figure 1.
Changes in the appearance, morphology, and lesion diameter of kiwifruit in the SHAM and CK groups induced by Botrytis cinerea at different days postinfection. (a) Changes in the morphological characteristics of kiwifruit at 0, 1, 3, and 5 d post infection (dpi). (b) Changes in the lesion diameter in kiwifruit in the SHAM and CK groups at different time points. '**' represents extremely significantly different (p < 0.01).
-
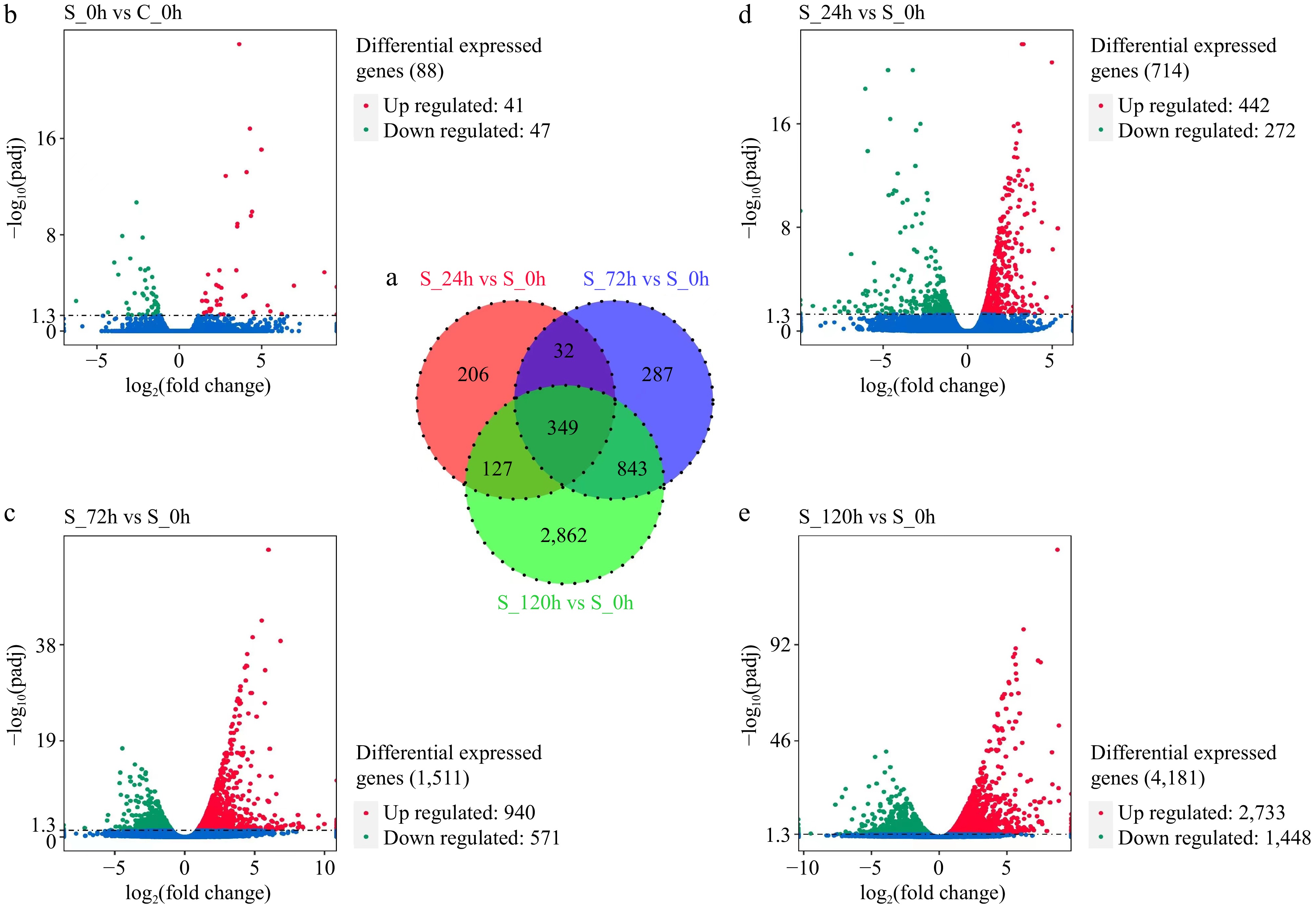
Figure 2.
(a) Venn diagram of differentially expressed genes (DEGs) between different groups (S_24h vs S_0h, S_72h vs S_0h, and S_120h vs S_0h). (b)–(e) Volcano plots of DEGs in different samples. Each dot represents a DEG. Red and green represent significantly up and downregulated DEGs.
-
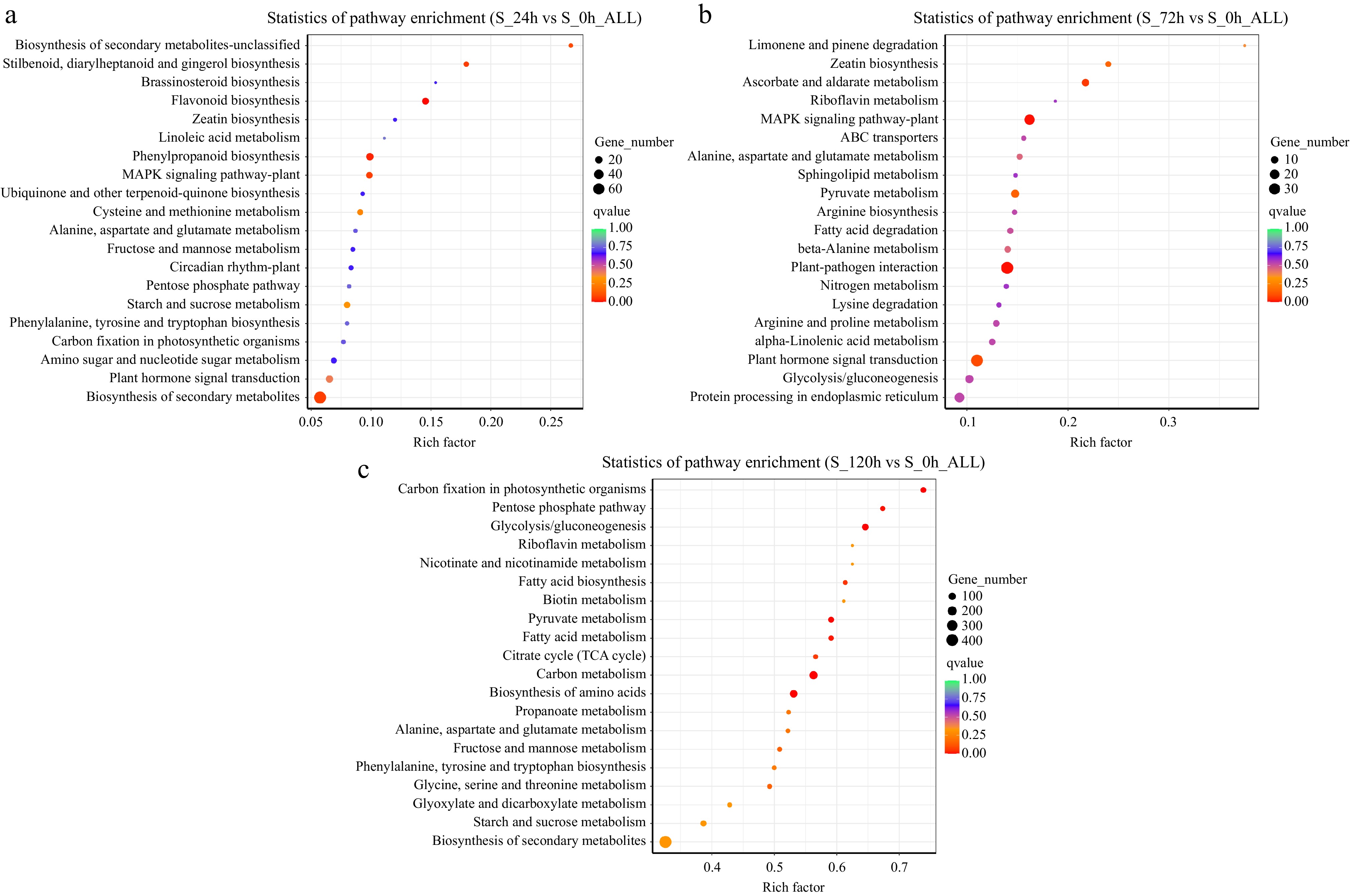
Figure 3.
(a)–(c) Analysis of enriched metabolic pathways in three groups. The top 20 most significant metabolic pathways (based on the p-values) are depicted, with each bubble representing a metabolic pathway. The size of the bubble represents the amount of metabolites in the pathway, as assessed through topological analysis. The color of the bubble represents the p-value of the enrichment analysis (i.e., −log10 p-value); the darker the color, the smaller the p value and the more significant the degree of enrichment.
-
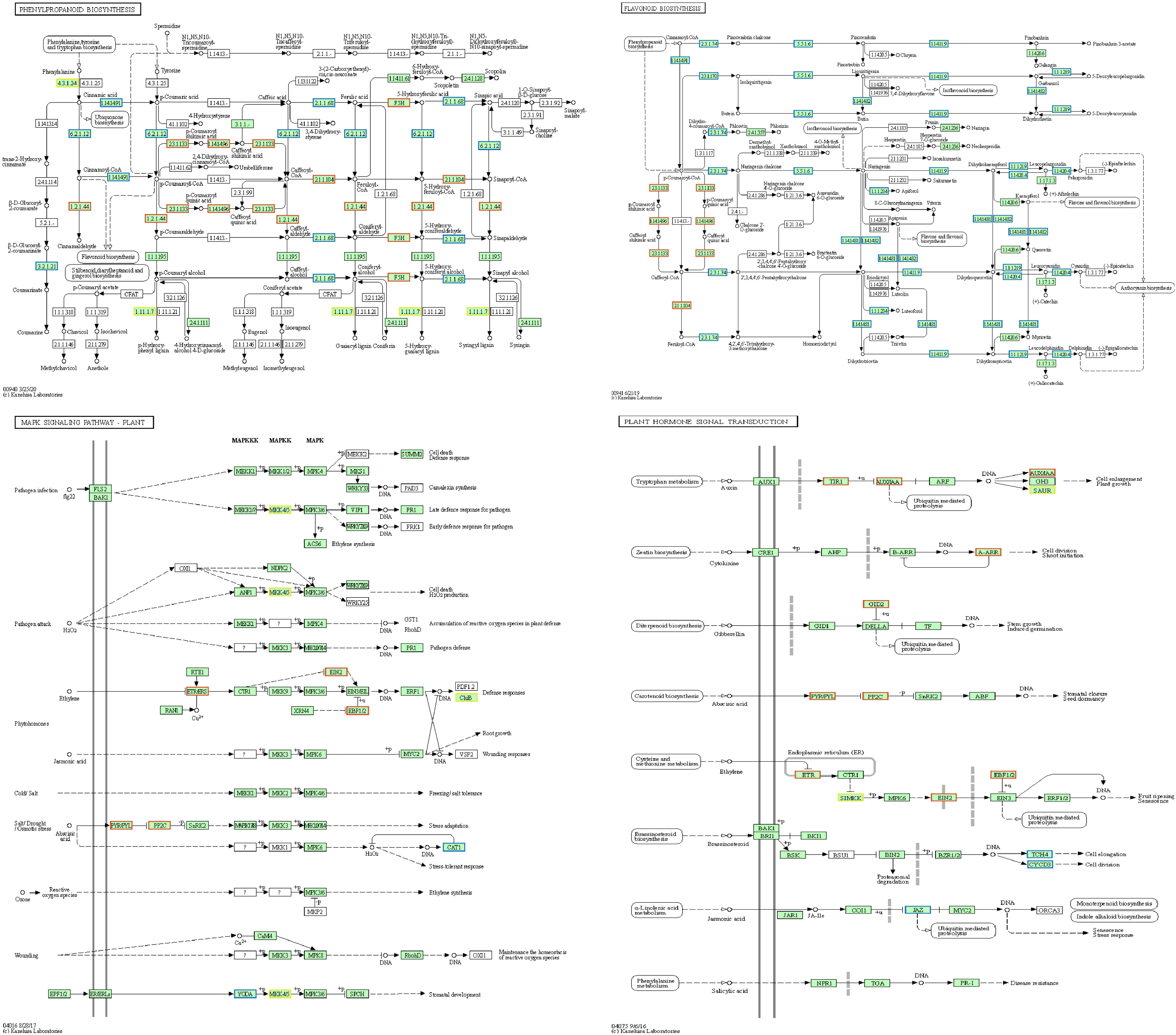
Figure 4.
Diagram showing the important KEGG pathways.
-
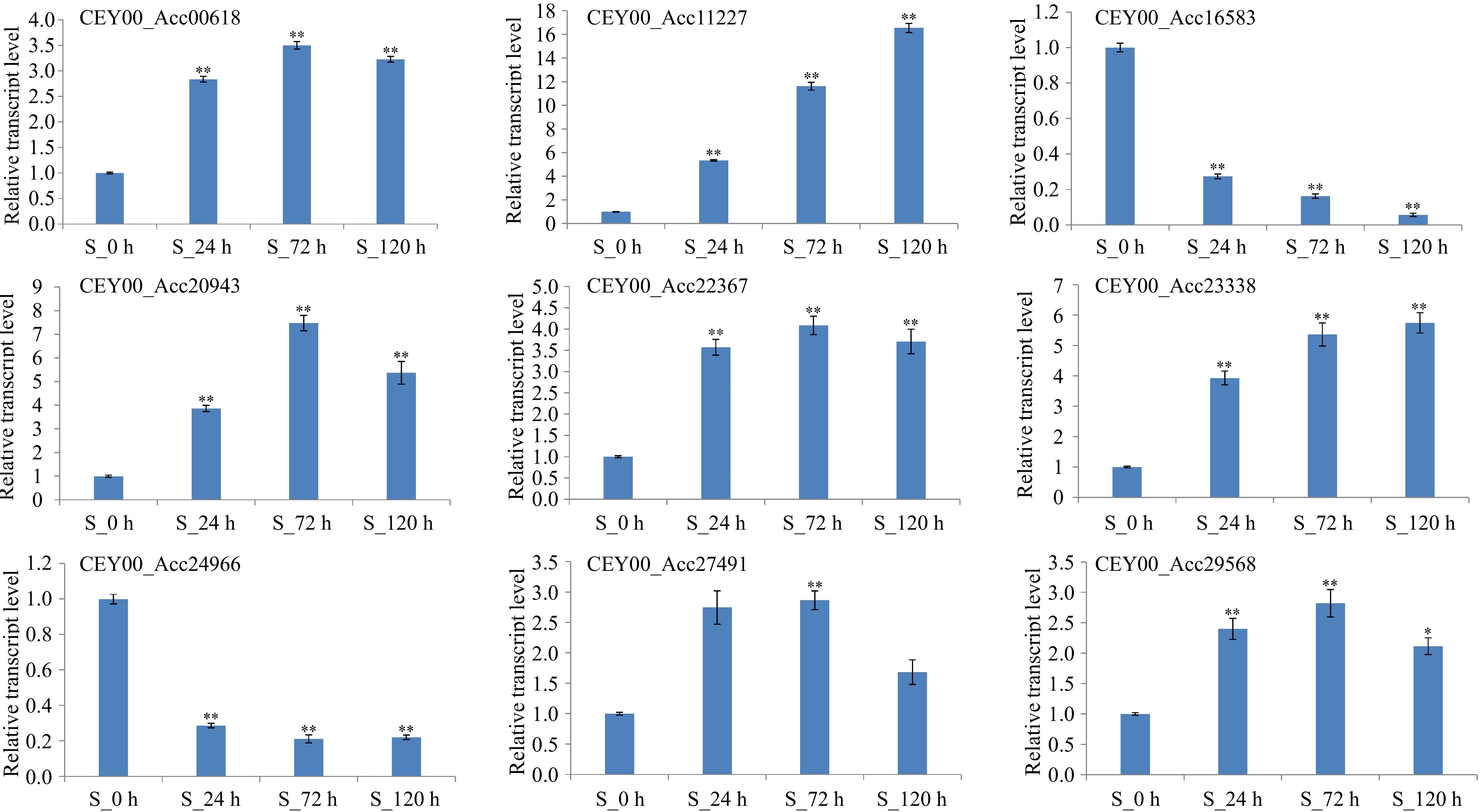
Figure 5.
qRT-PCR analysis of nine genes enriched for plant disease resistance pathways. CEY00_Acc00618: EIN3-binding F-box protein; CEY00_Acc11227: Caffeoyl-CoA O-methyltransferase; CEY00_Acc16583: Trans-cinnamate 4-monooxygenase; CEY00_Acc20943: Phenylalanine ammonia-lyase; CEY00_Acc22367: Mitogen-activated protein kinase; CEY00_Acc23338: Cytokinin dehydrogenase; CEY00_Acc24966: Chalcone synthase; CEY00_Acc27491: Ethylene receptor like; CEY00_Acc29568: Shikimate O-hydroxycinnamoyltransferase. '*' represents significantly different (p < 0.05), '**' represents extremely significantly different (p < 0.01).
-
Purpose Gene ID Primers (5'-3') qRT-PCR β-Actin F ACCGACATTTTCCTGCAACC R AGCAGCTGAGGTTGATCTGT qRT-PCR CEY00_Acc00618 F CGTTGGGTTTGTGCTCTGAAA R TCCTCAAACATGTCACCGGA qRT-PCR CEY00_Acc11227 F CCCATAATCGAAAAGGCCGG R TCACCAGATCGATCAGCCTC qRT-PCR CEY00_Acc16583 F CTTGAACCACCGCAACTTGA R CCTACGCATCTTCCTCCAGT qRT-PCR CEY00_Acc20943 F CTCCGGTGATCTGATCCCTCT R ATTCACTAGTGCCAGGCCTT qRT-PCR CEY00_Acc22367 F CCGAGACTTTGTTGCTTGCTC R CAGACCCTCCCGATTCTCTC qRT-PCR CEY00_Acc23338 F TTCCAACTCCTCCATCCCAC R GCCGATGATGGGTGAAGAAC qRT-PCR CEY00_Acc24966 F ACTGGTCGTCTGCTCTGAAA R GCCCCATCACTATCCGGTAA qRT-PCR CEY00_Acc27491 F TTGTTCAGTTGATGCAGGGCC R AAACAGTGCACCCGAGTTTC qRT-PCR CEY00_Acc29568 F ACCAGACTGCCCATTCATGA R GCACGGCTTTGTCAGATGAT Table 1.
Primers used in the present study.
-
Sample name Raw reads Raw bases Clean reads Clean bases Error rate Q20 Q30 GC content C_0h_1 47524076 7.16G 47391498 7.16G 0.03% 97.65% 93.24% 46.76% C_0h_2 46500794 7.01G 46365486 7G 0.03% 97.81% 93.62% 46.71% C_0h_3 40160680 6.05G 40052962 6.05G 0.03% 97.69% 93.27% 46.65% C_24h_1 42614752 6.42G 42488828 6.42G 0.03% 97.58% 93.05% 46.57% C_24h_2 43818432 6.6G 43697220 6.6G 0.03% 97.61% 93.09% 46.66% C_24h_3 39659046 5.98G 39568666 5.97G 0.03% 97.84% 93.61% 46.81% C_72h_1 39707836 5.98G 39601106 5.98G 0.03% 97.59% 93.07% 46.75% C_72h_2 39646984 5.98G 39546362 5.97G 0.03% 97.83% 93.63% 46.86% C_72h_3 40853092 6.16G 40733706 6.15G 0.03% 97.63% 93.18% 46.87% C_120h_1 38720330 5.83G 38616554 5.83G 0.03% 97.64% 93.17% 47.17% C_120h_2 45336366 6.83G 45202468 6.83G 0.03% 97.63% 93.15% 46.95% C_120h_3 42699226 6.43G 42577918 6.43G 0.03% 97.70% 93.31% 46.80% S_0h_1 47755908 7.2G 47622990 7.19G 0.03% 97.70% 93.31% 46.48% S_0h_2 45493394 6.85G 45362482 6.85G 0.03% 97.83% 93.63% 46.61% S_0h_3 42211600 6.36G 42099856 6.36G 0.03% 97.69% 93.28% 46.62% S_24h_1 44252586 6.67G 44112658 6.66G 0.03% 97.57% 93.04% 46.90% S_24h_2 38169744 5.75G 38062452 5.75G 0.03% 97.62% 93.15% 46.95% S_24h_3 38973972 5.87G 38878598 5.87G 0.03% 97.87% 93.72% 47.05% S_72h_1 44924206 6.77G 44725720 6.75G 0.03% 97.32% 92.49% 47.12% S_72h_2 41152860 6.2G 41023726 6.19G 0.03% 97.86% 93.72% 47.19% S_72h_3 40050140 6.04G 39942390 6.03G 0.03% 97.62% 93.14% 46.96% S_120h_1 37938536 5.72G 37839310 5.71G 0.03% 97.61% 93.12% 47.26% S_120h_2 42220850 6.36G 42087276 6.36G 0.03% 97.58% 93.15% 47.34% S_120h_3 42243486 6.36G 42114890 6.36G 0.03% 97.69% 93.37% 47.24% Table 2.
Statistics of sequencing data.
-
Sample name Total reads Total mapped Multiple mapped Uniquely mapped Read-1 Read-2 Readsmapto'+' Readsmapto'–' Non-splicereads Splicereads C_0h_1 47391498 44964944 (94.88%) 1713494 (3.62%) 43251450 (91.26%) 21625725 (45.63%) 21625725 (45.63%) 21625725 (45.63%) 21625725 (45.63%) 26448459 (55.81%) 16802991 (35.46%) C_0h_2 46365486 44059000 (95.03%) 1625532 (3.51%) 42433468 (91.52%) 21216734 (45.76%) 21216734 (45.76%) 21216734 (45.76%) 21216734 (45.76%) 25835515 (55.72%) 16597953 (35.8%) C_0h_3 40052962 38138198 (95.22%) 1437330 (3.59%) 36700868 (91.63%) 18350434 (45.82%) 18350434 (45.82%) 18350434 (45.82%) 18350434 (45.82%) 22123727 (55.24%) 14577141 (36.39%) C_24h_1 42488828 39228228 (92.33%) 1541524 (3.63%) 37686704 (88.7%) 18843352 (44.35%) 18843352 (44.35%) 18843352 (44.35%) 18843352 (44.35%) 23334117 (54.92%) 14352587 (33.78%) C_24h_2 43697220 41131072 (94.13%) 1626420 (3.72%) 39504652 (90.41%) 19752326 (45.2%) 19752326 (45.2%) 19752326 (45.2%) 19752326 (45.2%) 23913823 (54.73%) 15590829 (35.68%) C_24h_3 39568666 36938290 (93.35%) 1450522 (3.67%) 35487768 (89.69%) 17743884 (44.84%) 17743884 (44.84%) 17743884 (44.84%) 17743884 (44.84%) 21343865 (53.94%) 14143903 (35.75%) C_72h_1 39601106 37270004 (94.11%) 1464402 (3.7%) 35805602 (90.42%) 17902801 (45.21%) 17902801 (45.21%) 17902801 (45.21%) 17902801 (45.21%) 21764204 (54.96%) 14041398 (35.46%) C_72h_2 39546362 37400570 (94.57%) 1504068 (3.8%) 35896502 (90.77%) 17948251 (45.39%) 17948251 (45.39%) 17948251 (45.39%) 17948251 (45.39%) 22088098 (55.85%) 13808404 (34.92%) C_72h_3 40733706 38245790 (93.89%) 1521084 (3.73%) 36724706 (90.16%) 18362353 (45.08%) 18362353 (45.08%) 18362353 (45.08%) 18362353 (45.08%) 22622406 (55.54%) 14102300 (34.62%) C_120h_1 38616554 33816150 (87.57%) 1574710 (4.08%) 32241440 (83.49%) 16120720 (41.75%) 16120720 (41.75%) 16120720 (41.75%) 16120720 (41.75%) 19925731 (51.6%) 12315709 (31.89%) C_120h_2 45202468 41753966 (92.37%) 1695002 (3.75%) 40058964 (88.62%) 20029482 (44.31%) 20029482 (44.31%) 20029482 (44.31%) 20029482 (44.31%) 24363679 (53.9%) 15695285 (34.72%) C_120h_3 42577918 39318158 (92.34%) 1846690 (4.34%) 37471468 (88.01%) 18735734 (44%) 18735734 (44%) 18735734 (44%) 18735734 (44%) 22576173 (53.02%) 14895295 (34.98%) S_0h_1 47622990 43144188 (90.6%) 1804166 (3.79%) 41340022 (86.81%) 20670011 (43.4%) 20670011 (43.4%) 20670011 (43.4%) 20670011 (43.4%) 25591719 (53.74%) 15748303 (33.07%) S_0h_2 45362482 41979524 (92.54%) 1554640 (3.43%) 40424884 (89.12%) 20212442 (44.56%) 20212442 (44.56%) 20212442 (44.56%) 20212442 (44.56%) 24638686 (54.32%) 15786198 (34.8%) S_0h_3 42099856 39315626 (93.39%) 1510886 (3.59%) 37804740 (89.8%) 18902370 (44.9%) 18902370 (44.9%) 18902370 (44.9%) 18902370 (44.9%) 22678429 (53.87%) 15126311 (35.93%) S_24h_1 44112658 41846144 (94.86%) 1662310 (3.77%) 40183834 (91.09%) 20091917 (45.55%) 20091917 (45.55%) 20091917 (45.55%) 20091917 (45.55%) 24359883 (55.22%) 15823951 (35.87%) S_24h_2 38062452 36153600 (94.98%) 1412136 (3.71%) 34741464 (91.27%) 17370732 (45.64%) 17370732 (45.64%) 17370732 (45.64%) 17370732 (45.64%) 21234325 (55.79%) 13507139 (35.49%) S_24h_3 38878598 36883350 (94.87%) 1465624 (3.77%) 35417726 (91.1%) 17708863 (45.55%) 17708863 (45.55%) 17708863 (45.55%) 17708863 (45.55%) 21502098 (55.31%) 13915628 (35.79%) S_72h_1 44725720 41128742 (91.96%) 1876526 (4.2%) 39252216 (87.76%) 19626108 (43.88%) 19626108 (43.88%) 19626108 (43.88%) 19626108 (43.88%) 24421759 (54.6%) 14830457 (33.16%) S_72h_2 41023726 38514036 (93.88%) 1702480 (4.15%) 36811556 (89.73%) 18405778 (44.87%) 18405778 (44.87%) 18405778 (44.87%) 18405778 (44.87%) 22568830 (55.01%) 14242726 (34.72%) S_72h_3 39942390 37195056 (93.12%) 1558874 (3.9%) 35636182 (89.22%) 17818091 (44.61%) 17818091 (44.61%) 17818091 (44.61%) 17818091 (44.61%) 21984331 (55.04%) 13651851 (34.18%) S_120h_1 37839310 34998938 (92.49%) 1566774 (4.14%) 33432164 (88.35%) 16716082 (44.18%) 16716082 (44.18%) 16716082 (44.18%) 16716082 (44.18%) 20896975 (55.23%) 12535189 (33.13%) S_120h_2 42087276 38673976 (91.89%) 1863200 (4.43%) 36810776 (87.46%) 18405388 (43.73%) 18405388 (43.73%) 18405388 (43.73%) 18405388 (43.73%) 23284707 (55.32%) 13526069 (32.14%) S_120h_3 42114890 38897188 (92.36%) 1769032 (4.2%) 37128156 (88.16%) 18564078 (44.08%) 18564078 (44.08%) 18564078 (44.08%) 18564078 (44.08%) 23428900 (55.63%) 13699256 (32.53%) Table 3.
Comparison of rReads and reference sequences.
Figures
(5)
Tables
(3)