-
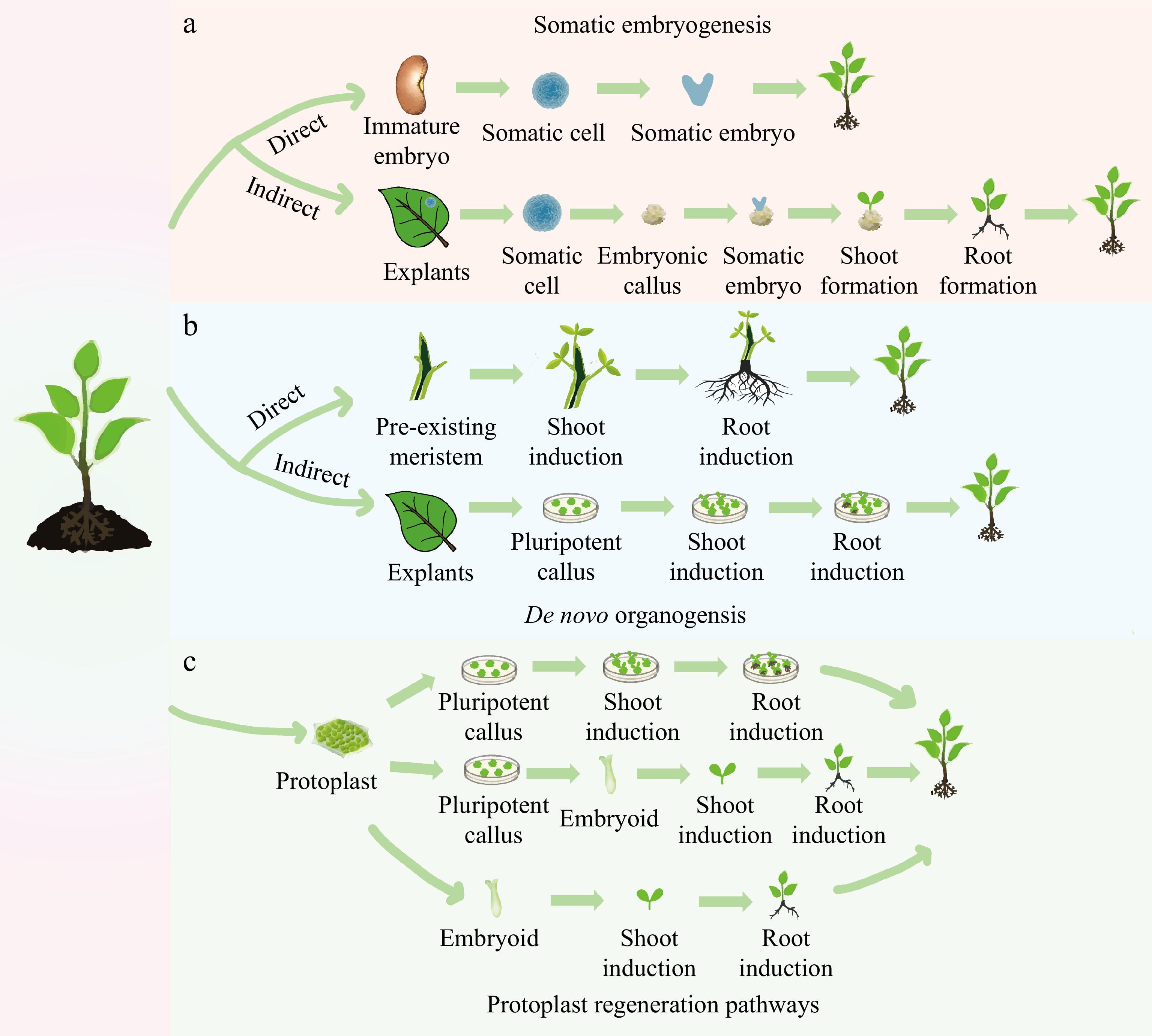
Figure 1.
Different pathways of plant regeneration. (a) Somatic embryogenesis: in the direct pathway, a somatic cell originating from an immature embryo is induced to form a somatic embryo, which then drives the development of the entire plant. In the indirect pathway, the explant is induced to initiate an embryonic callus, on which somatic embryos are formed. These embryos subsequently develop into shoots and roots. (b) De novo organogenesis: in the direct pathway, shoots and roots are induced directly on the stem with pre-existing meristems. In the indirect pathway, a pluripotent callus is produced around the wound in a leaf explant, with the formation of shoots and roots being subsequently induced. (c) Three methods of protoplast regeneration: one method involves the formation of a callus by protoplast differentiation, which is then induced to form shoots and roots, eventually differentiating into a plant. Another method involves differentiation from a protoplast to form a callus, followed by differentiation from a callus to an embryoid, and finally, the development of the whole plant. A direct method involves differentiating from a protoplast into an embryoid, which then develops into the whole plant.
-
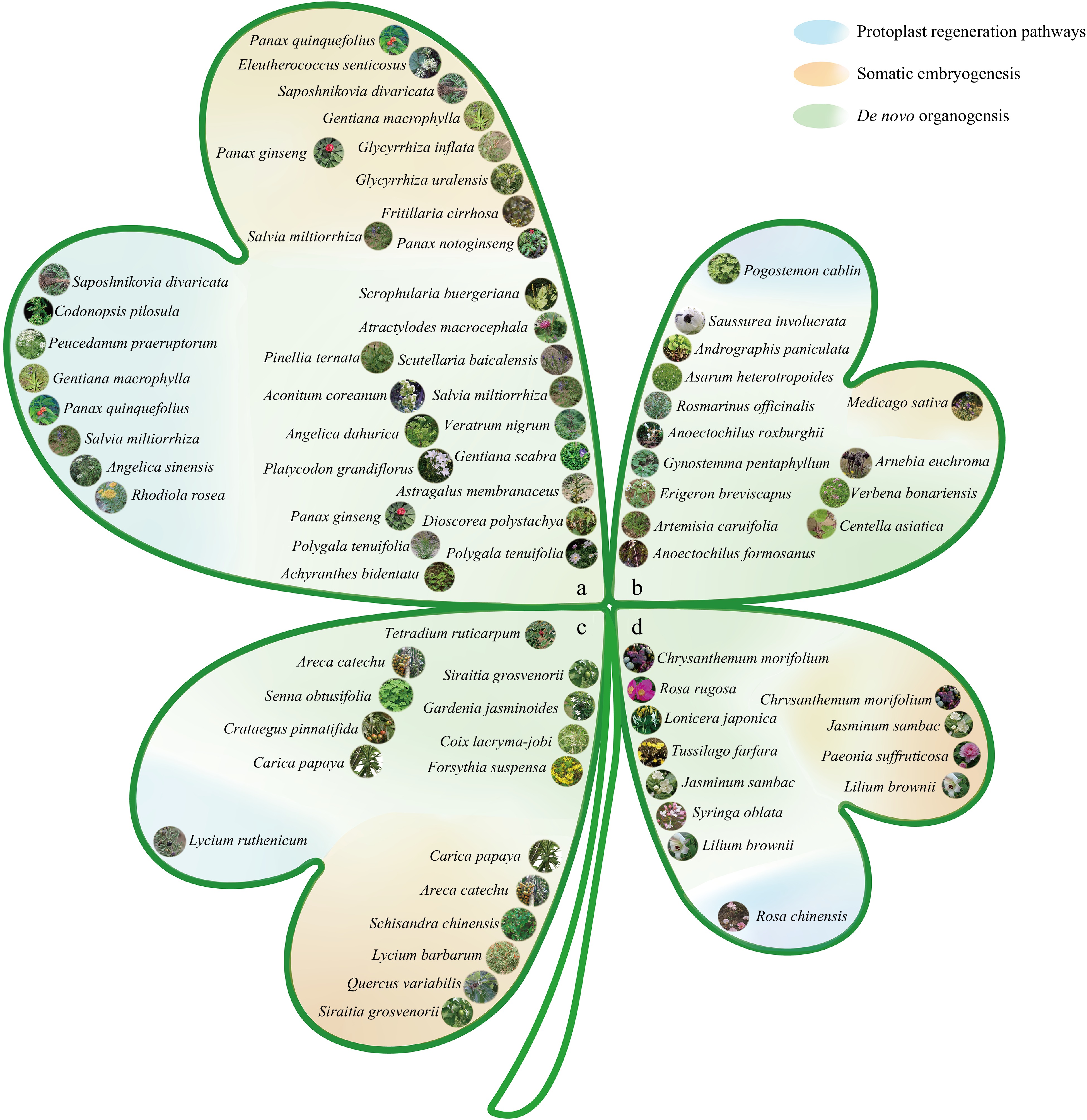
Figure 2.
Classification map of medicinal plants based on different medicinal parts and regeneration pathways. (a) Roots and rhizomes: arrangement of medicinal plants whose medicinal parts are roots and rhizomes. (b) Whole grass: arrangement of medicinal plants whose medicinal parts are the entire grass. (c) Fruits and seeds: arrangement of medicinal plants whose medicinal parts are fruits and seeds. (d) Flowers: arrangement of medicinal plants whose medicinal parts are flowers. Regeneration routes: the map is divided into three categories on the basis of different regeneration routes: the blue section represents medicinal plants regenerated through protoplast regeneration; the orange section represents medicinal plants regenerated through somatic embryogenesis; and the green section represents medicinal plants regenerated through de novo organogenesis. Note: All images are from Flora of China[13].
-
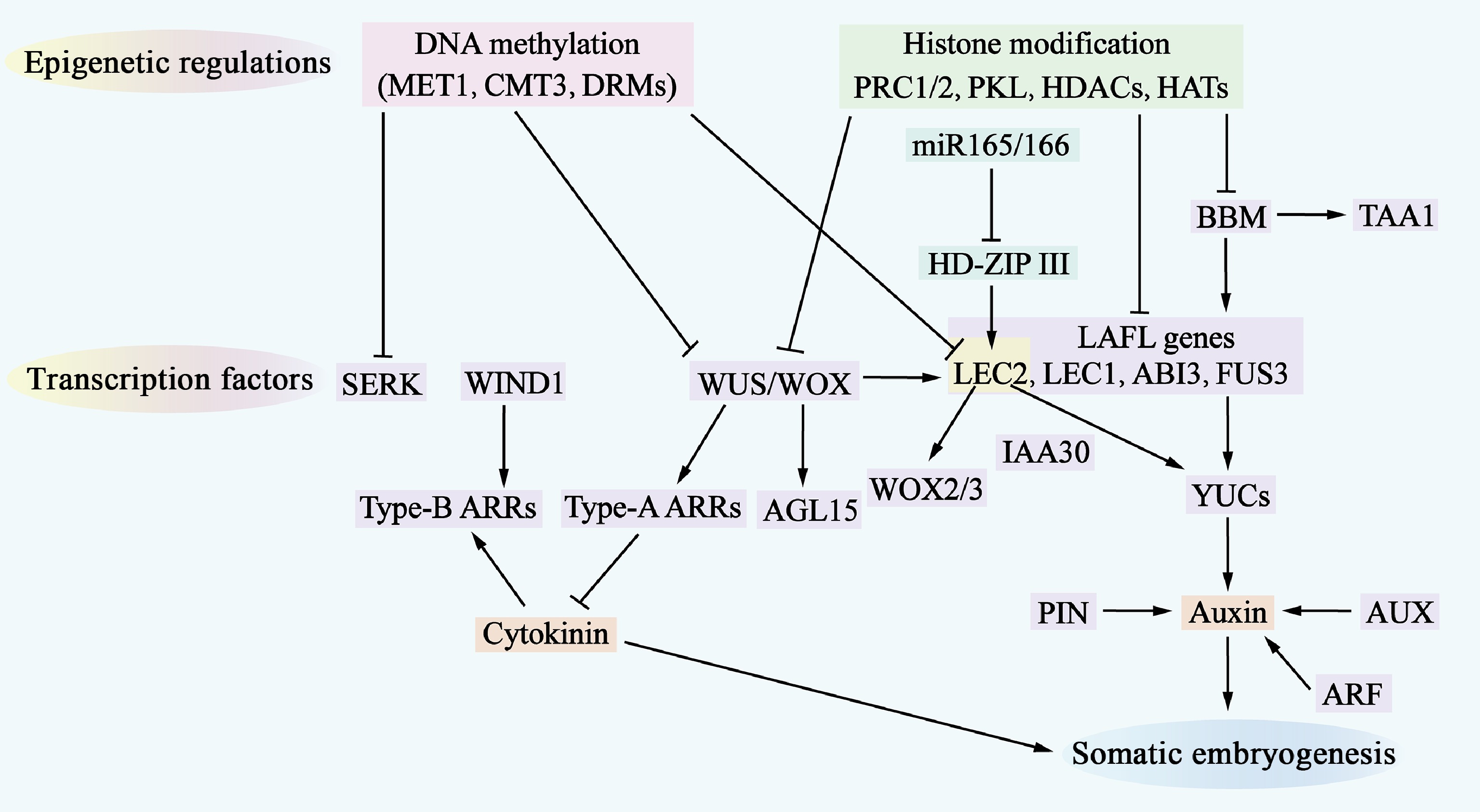
Figure 3.
Molecular mechanisms of somatic embryogenesis. The process of somatic embryogenesis is influenced by epigenetic regulation, transcription factors, and hormone signaling pathways. Epigenetic regulation, which includes DNA methylation (indicated by a pink shadow) and histone modifications (indicated by a green shadow), represses the access of transcription factors to gene-promoter regions, thereby inhibiting the expression of genes involved in somatic embryogenesis. Numerous transcription factors (indicated by a purple shadow) are involved in this regulatory network, where they also regulate each other and activate downstream auxin and cytokinin (CK) signaling pathways. Additionally, miR-165/-166 (indicated by a cyan shadow) are involved in regulating somatic embryogenesis.
-
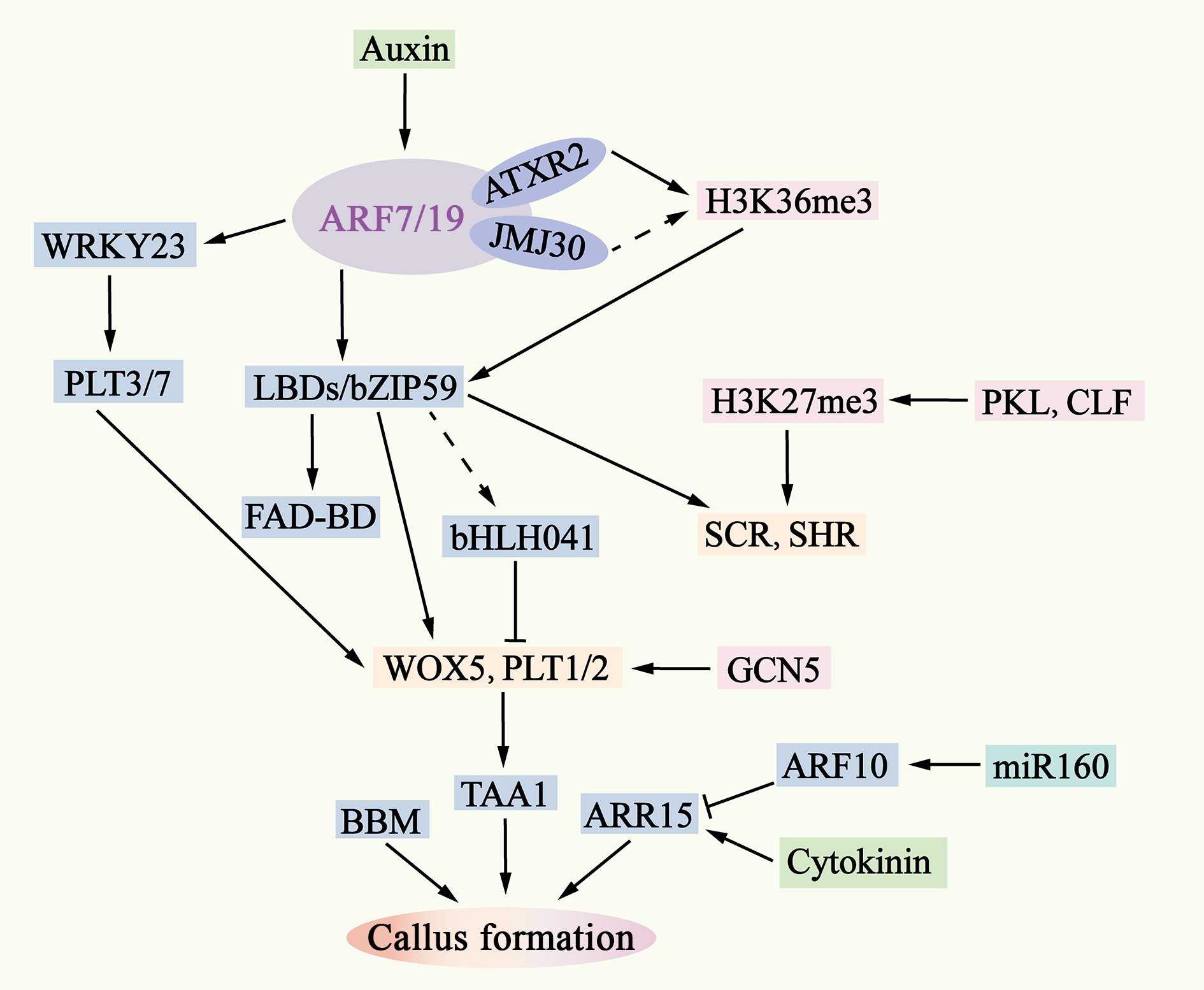
Figure 4.
Molecular mechanisms of pluripotent callus formation. The ability of auxin to activate ARF7/19 is also regulated by epigenetic factors such as AXTR2 and JMJ30. Downstream of ARF7/19, LBD-bZIP59 and WRKY23 activate the expression of genes such as WOX5, PLT1/2, SHR, and SCR by removing bHLH041 and PLT3/7. The interaction between WOX5 and PLT1/2 enhances the expression of TAA1, leading to an increase in endogenous auxin levels in the callus and inducing the formation of pluripotent calli. The removal is indicated by dashed lines.
-
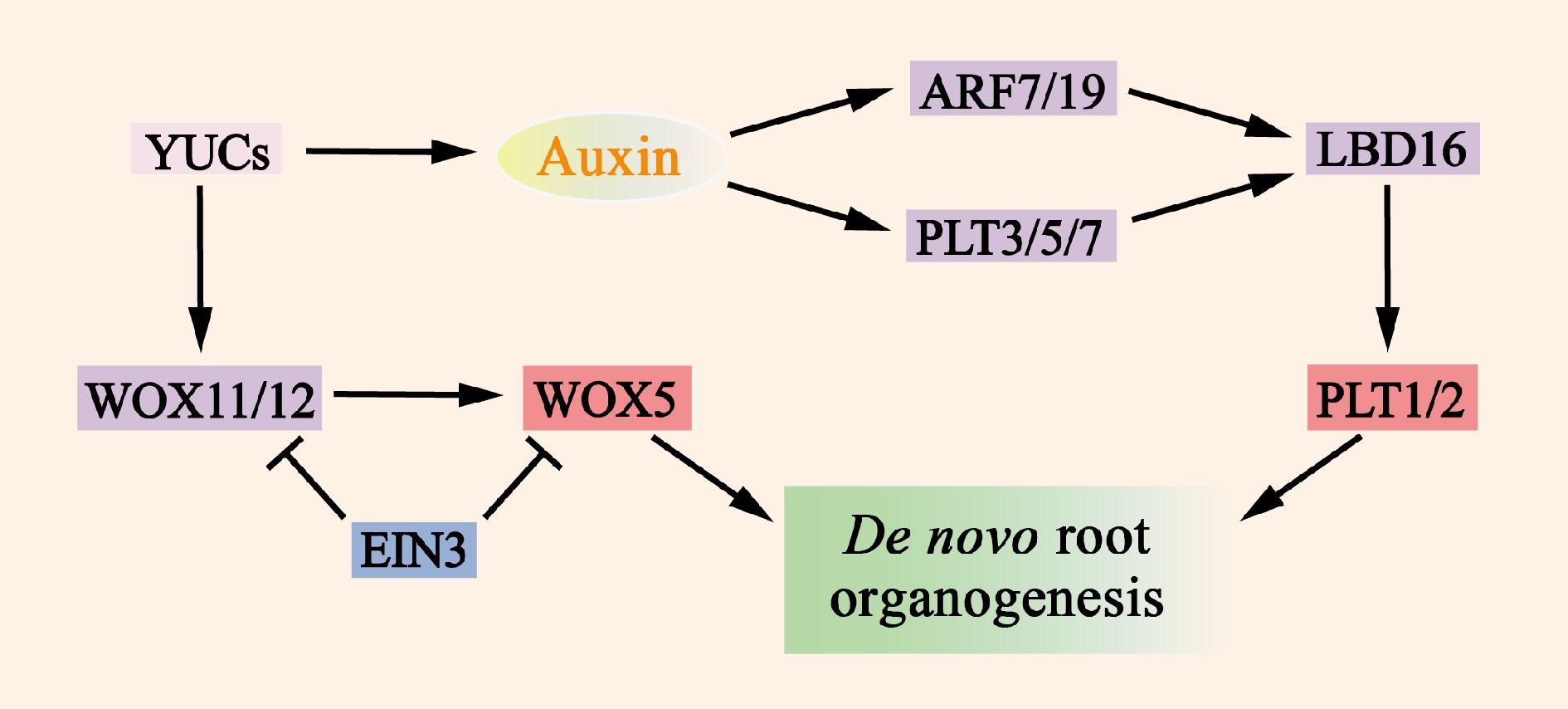
Figure 5.
Molecular mechanisms of de novo root organogenesis. Auxin, which is mediated by YUC, serves as a key regulatory factor that activates the expression of WOX11/12, ARF7/19, and PLT3/5/7. The translation products of these genes then directly or indirectly promote the expression of WOX5 and PLT1/2, which in turn induce the formation of de novo root organogenesis. The transcription factor EIN3 significantly reduces the frequency of new root organs by inhibiting the transcription of WOX11 and WOX5.
-
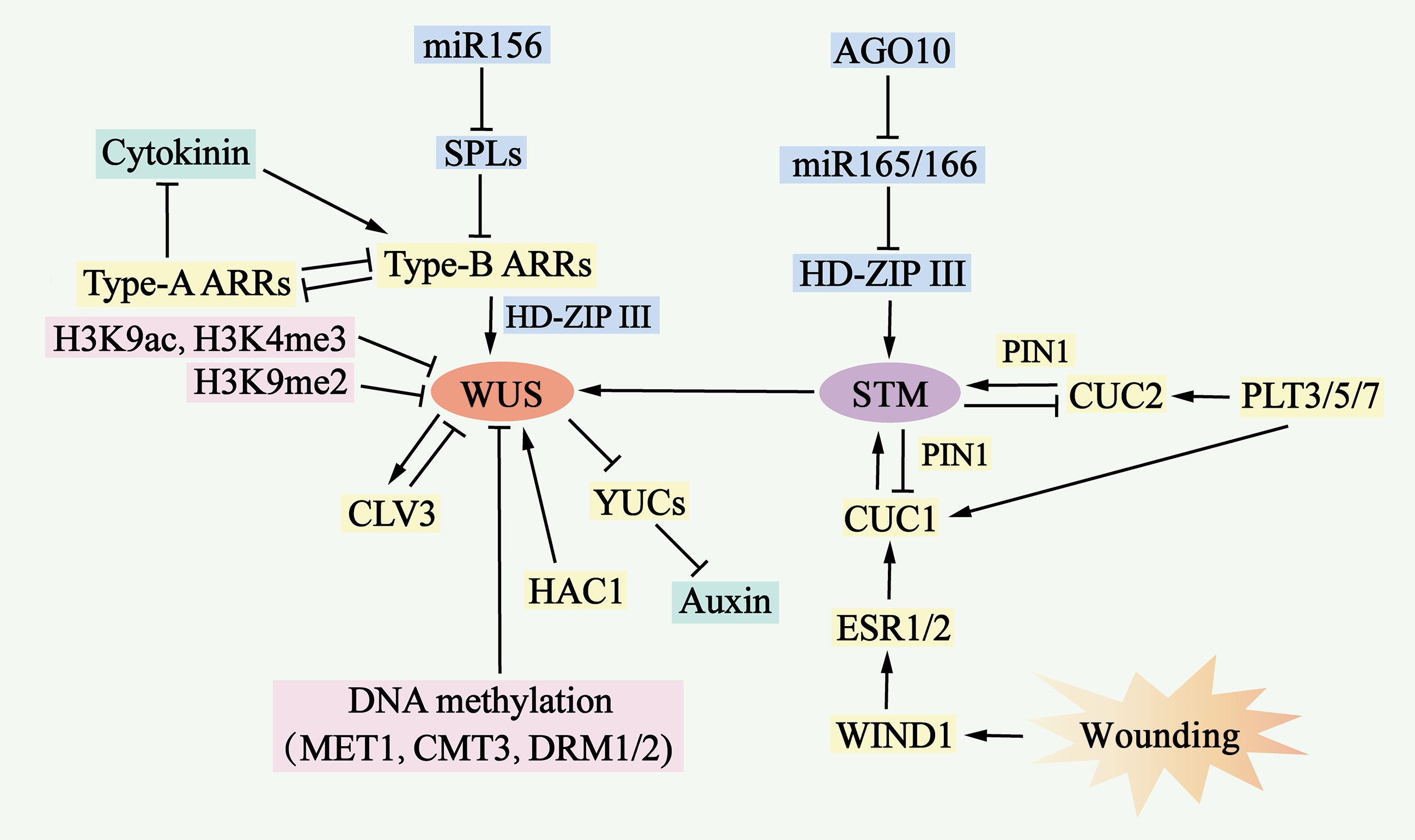
Figure 6.
Molecular mechanisms of de novo shoot organogenesis. During the process of de novo shoot organogenesis, two pathways, the WUS-CLV3 pathway and the STM-CUC pathway, establish negative feedback loops and play critical regulatory roles. The WUS-CLV3 pathway is regulated primarily by DNA methylation, histone modification, and hormone signaling. Cytokinin (CK) activates the expression of type B ARRs, which in turn stimulates WUS expression, whereas type B ARRs repress YUC-mediated auxin biosynthesis. In the STM-CUC pathway, STM expression is promoted by CUC1 and CUC2, both of which are upregulated by PLT3/5/7, ESR1, ESR2, WIND1, and PIN1. Moreover, WUS and STM interact directly to activate CLV3 expression, suggesting that the two pathways converge and coordinate to control shoot regeneration.
Figures
(6)
Tables
(0)