-
Medicinal plant resources constitute the cornerstone of the sustained and healthy development of the traditional Chinese medicine (TCM) industry, as they not only treat diseases but also strengthen the human body's immunity and prevent the occurrence of diseases[1]. As an important country for the global production and use of medicinal plants[2], China has more than 13,000 species of medicinal plants, of which more than 200 commonly used Chinese herbal medicine have been planted on a large scale[3]. However, with the modernization of TCM and the expansion of the Chinese herbal medicine industry, the demand for Chinese herbal medicine has also increased annually. The rapid development of TCM, diet, health care, cosmetics, and other industries has led to the over-exploitation and large-scale use of medicinal plant resources, and many rare medicinal plant resources are facing a depletion crisis[2]. Therefore, the establishment of a regeneration system for medicinal plants is highly important to protect these valuable resources, especially endangered medicinal plants.
To ensure the safety and efficacy of clinical drugs, it is essential to prioritize the development of high-quality Chinese herbal medicine. Biological breeding technology, especially the application of gene editing technology, is expected to lead the innovation of molecular breeding technology in TCM. Medicinal plant regeneration technology is key to achieve this goal[3]. Currently, the regeneration of medicinal plants is focused mainly on the application level, and systematic investigations and in-depth considerations of regeneration mechanisms and future development are still insufficient. The aim of this study was to elaborate on the regeneration pathways, influencing factors and molecular mechanisms of medicinal plants to provide new perspectives and ideas for the sustainable utilization of Chinese herbal medicine resources.
-
To achieve totipotency, differentiated cells must first undergo dedifferentiation, then redifferentiation. Dedifferentiated plant explants form healing tissues, which are transferred to differentiation medium, where morphogenesis is completed, resulting in shoot and root structures, which mature into complete plants.[4]. In vitro tissue culture of medicinal plants, plant regeneration is achieved mainly through somatic embryogenesis (SE), de novo organogenesis, and protoplast regeneration (Fig. 1).
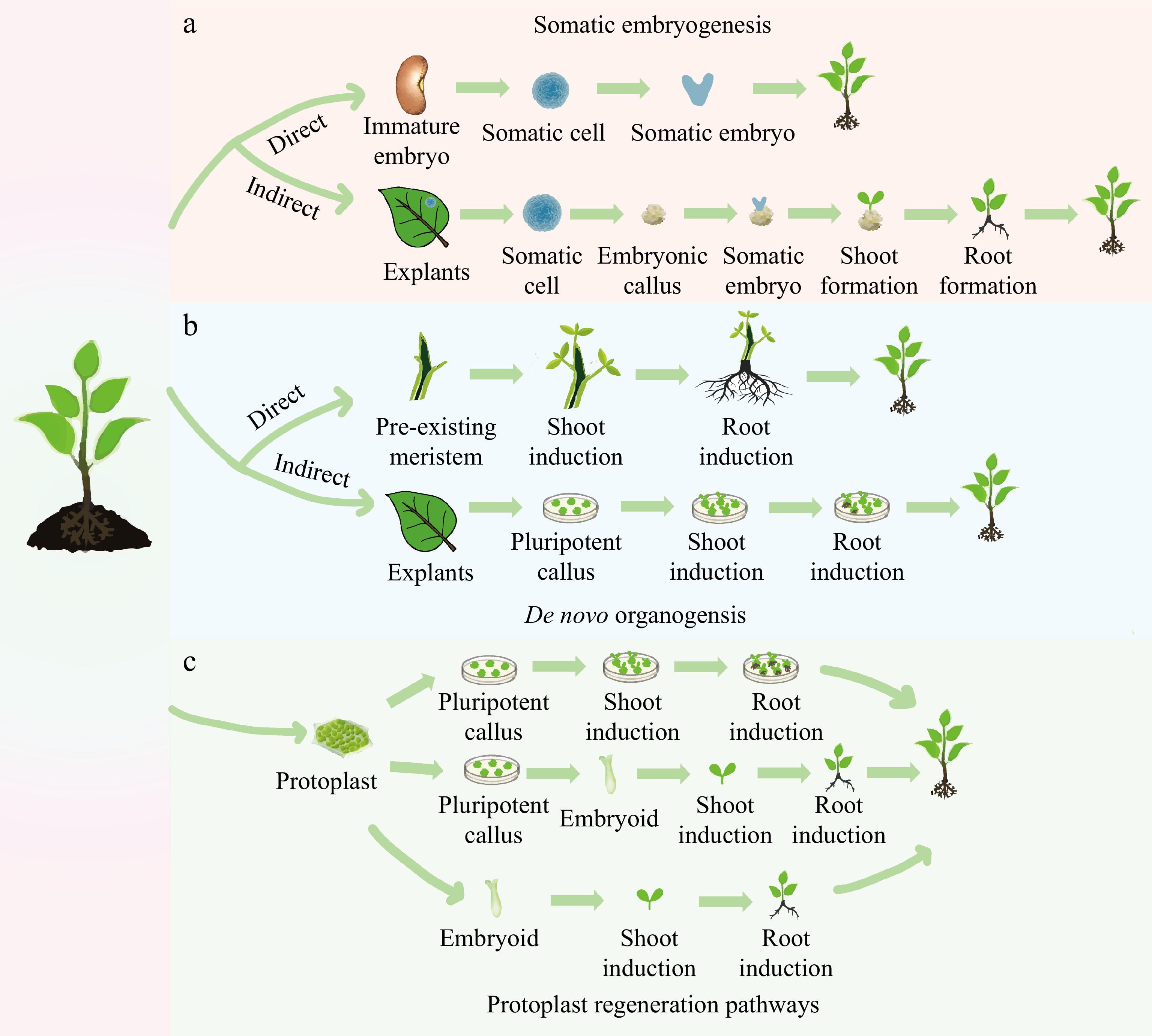
Figure 1.
Different pathways of plant regeneration. (a) Somatic embryogenesis: in the direct pathway, a somatic cell originating from an immature embryo is induced to form a somatic embryo, which then drives the development of the entire plant. In the indirect pathway, the explant is induced to initiate an embryonic callus, on which somatic embryos are formed. These embryos subsequently develop into shoots and roots. (b) De novo organogenesis: in the direct pathway, shoots and roots are induced directly on the stem with pre-existing meristems. In the indirect pathway, a pluripotent callus is produced around the wound in a leaf explant, with the formation of shoots and roots being subsequently induced. (c) Three methods of protoplast regeneration: one method involves the formation of a callus by protoplast differentiation, which is then induced to form shoots and roots, eventually differentiating into a plant. Another method involves differentiation from a protoplast to form a callus, followed by differentiation from a callus to an embryoid, and finally, the development of the whole plant. A direct method involves differentiating from a protoplast into an embryoid, which then develops into the whole plant.
SE pathway
-
SE is an important plant regeneration technique that allows somatic cells to undergo a series of developmental changes that culminate in the formation of embryo-like structures. This process reveals the totipotency of plant cells through embryogenic calli; they can dedifferentiate into embryonic stem cells and redifferentiate into complete plants[5,6]. This change in cell fate is usually realized under specific stress conditions, hormone induction (such as auxin) or gene expression modification, and has great potential for plant reproduction, genetic transformation and the protection of rare and endangered species[7−9]. The formation of somatic embryos can be achieved via two routes: direct induction from individual somatic cells or indirect induction through the healing of embryonic tissue[10] (Figs 1a & 2). The direct route involves the induction of embryoid formation from the epidermis, subepidermis, young embryos, cells in suspension culture, and protoplasts of explants. For example, Glycyrrhiza uralensis hypocotyls can be used as explants to directly induce embryoid formation on MS medium and successfully culture regenerated plants[11]. The indirect pathway is more common and begins with embryonic healing tissue induction, followed by the formation of pre-embryonic masses on the surface or inside of healing tissues, which then develop into somatic embryos. Under the right conditions, these embryos are capable of further developing into complete plants with roots and shoots[12].
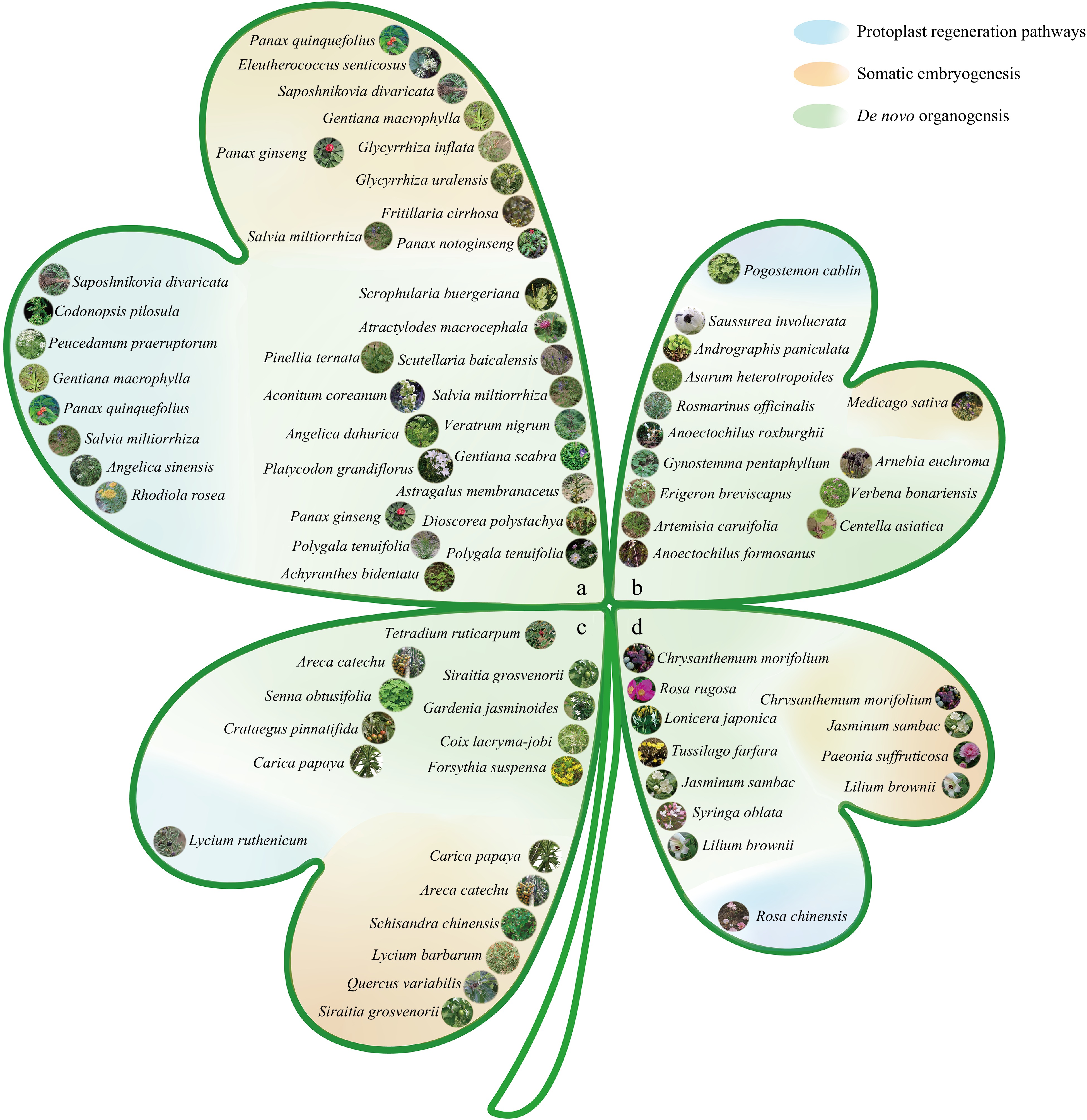
Figure 2.
Classification map of medicinal plants based on different medicinal parts and regeneration pathways. (a) Roots and rhizomes: arrangement of medicinal plants whose medicinal parts are roots and rhizomes. (b) Whole grass: arrangement of medicinal plants whose medicinal parts are the entire grass. (c) Fruits and seeds: arrangement of medicinal plants whose medicinal parts are fruits and seeds. (d) Flowers: arrangement of medicinal plants whose medicinal parts are flowers. Regeneration routes: the map is divided into three categories on the basis of different regeneration routes: the blue section represents medicinal plants regenerated through protoplast regeneration; the orange section represents medicinal plants regenerated through somatic embryogenesis; and the green section represents medicinal plants regenerated through de novo organogenesis. Note: All images are from Flora of China[13].
SE in medicinal plants usually takes place in an indirect manner, where embryonic healing tissues are induced via different explants, which then differentiate to form embryoid bodies and eventually develop into plants[8] (Figs 1a & 2). For example, the regeneration of medicinal plants such as Picrorhiza kurroa[14], Panax notoginseng[15], and Eleutherococcus senticosus[16] has been achieved in this way. In addition, the hairy roots of plants such as Scrophularia buergeriana[17], and Panax quinquefolius[18] can also induce the formation of healing tissues in specific culture medium, subsequently forming embryos and regenerating new individuals.
Somatic embryos produced via direct and indirect routes are morphologically similar but differ in culture time, susceptibility to genetic variation, and plant regeneration potential. Although the indirect route requires a longer culture time and is more susceptible to genetic variation, it is capable of producing large amounts of healing tissue, which increases the regeneration potential of plants[19,20]. In contrast, the direct route is more efficient at producing a limited number of regenerated plants. Therefore, the direct route is more appropriate when the goal is to regenerate a specific number of plants rapidly. However, the indirect route provides a better alternative for species from which explants are difficult to obtain or for which many regenerated plants are needed[21].
De novo organogenesis pathway
-
The organogenesis pathway is a regenerative process that does not depend on somatic embryos and is achieved through the differentiation of meristematic tissue centers, demonstrating the pluripotency of plant cells[22]. This regeneration mechanism allows plants to regenerate de novo root and/or de novo shoot in vitro or from damaged organs, a common phenomenon in nature. The processes of regenerating de novo shoots and de novo roots are referred to as de novo shoot organogenesis and de novo root organogenesis, respectively[23]. Although de novo organogenesis is simple to induce, with a high induction rate, it may lead to the formation of chimeras. Like SE, de novo organogenesis can be initiated directly or indirectly (Fig. 1b).
The direct organogenesis pathway facilitates the rapid generation of regenerating plants from tissue organs such as stem tips, stems, metamorphic stems, and leaves of medicinal plants, eliminating the healing tissue induction stage and thus shortening the regeneration cycle[2] (Fig. 1b). This pathway is widely used in medicinal plants in China, such as Andrographis paniculata[24] Platycodon grandiflorus[25], Dendrobium nobile[26], and Schisandra chinensis[27], which are not only in high market demand but also maintain genetic stability during the culture process.
The indirect organogenesis pathway involves a distinct healing tissue stage during regenerating plant culture; this pathway involves a wider selection of explants, including anthers, leaves, stem segments, petioles, and roots. The indirect pathway has relatively high amplification and transformation rates, but there are differences in the morphogenetic capacity of explants from different parts of the plant (Figs 1b & 2). For example, the flowering branches and inflorescences of Artemisia caruifolia are more effective than the leaves in healing tissue induction and shoot differentiation[28]. Previous studies have shown that the healing capacity of leaves from Ligusticum sinense is greater than that of stem segments and roots, whereas the healing capacity of leaves is similar to that of stem segments; roots are weaker in terms of shoot differentiation and rooting capacity. In general, the ability of plant spindles to differentiate and regenerate gradually decreases from top to bottom[29].
In recent years, China has made remarkable progress in cultivating regenerated plant species through the organogenesis pathway, covering not only bulk medicinal plants such as the Asteraceae, Liliaceae, Labiatae, Araliaceae, Ranunculaceae, and Dioscoreaceae families but also slow-growing, endangered or progressively endangered medicinal plants such as Dendrobium officinale, Neopicrorhiza scrophulariiflora, Coptis teeta and Ginkgo biloba. At present, regeneration systems have been established for more than 200 medicinal plants[2] (Fig. 2).
The essential difference between de novo organogenesis and SE is that the former does not involve the formation of somatic embryos. Both pathways involve direct and indirect methods of regeneration, but the indirect methods differ with respect to healing tissue characteristics. SE produces totipotent embryonic healing tissues, whereas de novo organogenesis induces pluripotent nonembryonic healing tissues[30,31] (Fig. 1a & b). Indirect de novo organogenesis may lead to genetic instability and somatic cell asexual lineage variation[32]. Direct organogenesis is a time-saving method but is not suitable for transgenic research because of the possibility of chimerism[33]. The development of somatic cell embryos for certain organs or tissues that readily induce de novo organogenesis is challenging. Therefore, a combination of both pathways is sometimes used to increase the frequency of plant regeneration in a particular species, either in the commercial market or in scientific research.
Protoplast regeneration pathways
-
In addition to de novo organogenesis and SE, scientists have discovered the ability of individual cells to regenerate whole plants. As early as 1954, a method for single-cell healing tissue culture of Tagetes erecta was established[34]. Healing tissues in vitro from the phloem of Daucus carota roots were cultured, and single cells capable of undergoing multiple rounds of division and eventually developing into somatic embryos were obtained[35]. At present, plant protoplast regeneration culture and suspension culture techniques have been widely used for plants of several families, such as the Rosaceae, Boraginaceae, and Scrophulariaceae families, indicating great potential for application[36] (Fig. 2).
In China, protoplast culture technology has been successfully applied to the culture of a variety of medicinal plants, including Gentiana Macrophylla, Peucedanum praeruptorum, Saposhnikovia divaricata, Salvia miltiorrhiza, Cercospora asparagi, Lycium chinense, Rhodiola rosea, and Codonopsis pilosula (Fig. 2). Protoplasts are used for plant regeneration through three main pathways: the differentiation of protoplasts to form healing tissues that then redifferentiate into plants; the direct differentiation of protoplasts into embryos that then develop into plants; and the differentiation of protoplasts to form healing tissues that then redifferentiate into embryos and eventually develop into plants (Fig. 1c). For example, many embryonic protoplasts were cultured from calli induced by cutting P. peucedanum seedlings, and whole plants were ultimately obtained[37]. Protoplasts were isolated from the hypocotyl callus of C. pilosula, and embryoids were obtained via shallow liquid culture and subsequently developed into complete plants[38]. Tissue culture was used to obtain tissue culture-generated seedlings of Rhodiola sachalinensis. The leaves of R. sachalinensis tissue culture-generated seedlings were hydrolyzed by enzymes to obtain protoplasts, and calli were obtained via shallow liquid culture. The calli differentiate into adventitious shoots and then develop into complete plants[39]. In addition, protoplasts have been isolated from S. miltiorrhiza plants via leaf enzymatic hydrolysis, and protoplast regeneration systems for S. miltiorrhiza have been established by targeting one or more sites with the sgRNA-Cas9 ribonucleoprotein (RNP) complex or a plasmid carrying CRISPR/Cas9 system genes[40].
Notably, compared with other model plants and crops, research on the protoplasts of medicinal plants is still immature, and the technology needs to be improved. Therefore, further studies of regeneration culture, functional gene analysis and metabolite synthesis mechanisms of medicinal plant protoplasts are highly important to promote the sustainable and efficient development and utilization of medicinal plant resources.
-
During the in vitro regeneration culture of medicinal plants, the plant genotype is an important factor affecting regeneration capacity. There are significant differences in the response of plants of different genotypes to in vitro culture, a phenomenon that may be related to genetic differences and gene regulation[41]. For example, when stem segments were used as explants in S. miltiorrhiza f. alba and S. miltiorrhiza Bunge, 100% of S. miltiorrhiza f. alba stem segments developed shoots[42], whereas 85% of S. miltiorrhiza Bunge stem segments developed shoots (Supplementary Table S1)[43]. In addition, S. miltiorrhiza Bunge varieties from different regions, such as Shandong S. miltiorrhiza and Sichuan S. miltiorrhiza, present differences in regeneration systems, with Shandong S. miltiorrhiza having a relatively high shoot induction rate and Sichuan S. miltiorrhiza having a relatively high rate of healing tissue induction (Supplementary
Table S1)[44]. When cotyledons of Astragalus membranaceus were used as explants, healing tissues were obtained from 90% of the cotyledons on healing induction medium, with an optimal shoot generation rate of 90%[45], whereas cotyledons of A. membranaceus var. mongholicus were used as explants, the rate of healing tissue induction was 25% on the same healing induction medium, with no shoot differentiation[46]. In studies of the tissue culture of Taraxacum mongolicum, the cultures of different types of T. mongolicum differ under the same conditions. For example, Menggu T. mongolicum and Liaodong T. mongolicum have relatively high frequencies of callus induction, at 85.3% and 76%, respectively, whereas Taraxacum ohwianum, Taraxacum coreanum and Republic of Korea T. mongolicum have relatively low rates of callus formation and severe browning (Supplementary Table S1)[47].In studies of G. uralensis, the highest frequency of in vitro regeneration (up to 44.7%) was observed when hypocotyls were used as explants, whereas the number of clumped seedlings was greater when cotyledonary stem segments and leaf-bearing stem segments were used as explants[11]. These findings suggest that there are significant differences in healing tissue induction among different genotypes and that the selection of suitable organs and materials is crucial for improving the induction rate and perfecting regeneration systems for medicinal plants. In practical applications, the regeneration efficiency of specific genotypes can be improved by optimizing the medium composition and culture conditions. For example, when calli of A. membranaceus were induced, the application of activated carbon and TCM extracts in improved MS medium significantly increased the induction rate (Supplementary Table S1)[45]. In addition, research progress on the tissue culture of G. uralensis has shown that the use of different concentrations of plant growth regulators (PGRs) and nitrogen sources has significant effects on callus induction and growth (Supplementary Table S1).
Plant growth regulators
-
Exogenous hormones, especially PGRs, such as auxins and cytokinins, play key roles in plant SE and neoorganogenesis[48]. In vitro plant regeneration depends on the addition of exogenous hormones and the response to these hormones during tissue culture[49]. One study reported that the response of explants to PGRs occurs in three phases: first, the perception of phytohormone signals by explant cells, which induces dedifferentiation; second, specific cellular differentiation influenced by hormone homeostasis, which lays the groundwork for organ differentiation; and last, the completion of plant morphogenesis independent of exogenous hormones[50].
Growth factors are determinants of SE, among which 2,4-dichlorophenoxyacetic acid (2,4-D)[51], a synthetic auxins, is a strong inducer of healing tissues and is widely used in many species, especially cereal crops and medicinal plants. The concentration of 2,4-D has a significant effect on the formation of healing tissues, with low concentrations promoting the formation of embryonic healing tissues, whereas high concentrations may inhibit their formation. In medicinal plants, 5–10 μM 2,4-D is often used to induce somatic embryos, but prolonged accumulation may have toxic effects on cells and increase the risk of somatic mutations (Supplementary Tables S1 & S2)[52].
Other growth factors include indole-3-acetic acid (IAA), 1-naphthaleneacetic acid (NAA) and indole-3-butyric acid (IBA). NAA is often chosen for inducing roots differentiation and has a stronger effect than IAA and IBA do. However, the roots after NAA treatment are thicker and shorter and easier to break, whereas the roots after IAA treatment are weaker and readily degrade. In addition, the roots obtained by IBA treatment are more robust; therefore, many researchers have used a combination of NAA and IBA to optimize roots induction. For example, the roots induction rate of Angelica sinensis was highest at a hormone ratio of 2.0 mg/L IBA + 0.01 mg/L NAA, and the roots induction of Hemsleya chinensis was strongly induced at a hormone ratio of 2.0 mg/L 6-BA + 0.5 mg/L NAA + 1.0 mg/L IBA (Supplementary Tables S1 &S2)[53−55].
Cytokinins are also widely used PGRs in tissue culture, especially for de novo shoot induction and SE initiation. A proposed model of hormonal control of regeneration is widely used in the regeneration of explants. 6-BA, a commonly used cytokinin, is often used in conjunction with other hormones to induce callus differentiation and bulblet induction (Supplementary Tables S1 & S2).
In addition to auxins and cytokinins, other plant hormones, such as abscisic acid (ABA) and gibberellin (GA), also affect plant regeneration[56]. For example, GA promotes the germination and differentiation of immature embryos, whereas ABA and other hormones, such as oleuropein steroids and abscisic acid, also induce healing in some species. GA can substitute for auxins or cytokinins in the formation of healing tissues (Supplementary Tables S1 & S2)[57,58].
There are large differences in the phytohormone requirements for the regeneration of different genotypes and explants of medicinal plants; therefore, plant growth regulators suitable for different species need to be screened to achieve increased induction efficiency. The use of plant growth regulators is also widely used in the production of herbal medicines, but attention needs to be given to their effects on the effectiveness and safety of herbal medicines.
Environmental stresses
-
The ability of cells to initiate a regenerative program first needs to overcome specific tissue fate restrictions. Stress can effectively liberate cells from their fate constraints, resulting in the disruption or loss of intercellular communication. By dissecting and dissociating explant tissues and applying osmotic stress, cytoplasmic lysis, intercellular junction rupture, or cell death can be induced, resulting in the separation of living cells and the elimination of intercellular interactions. Isolated cells usually have thick cell walls and few or no intercellular junctions and exhibit cellular morphological features characteristic of stem cells, such as a large central nucleus and dense cytoplasm[6,59]. Owing to the loss of intercellular interactions, these cells may be excluded from fate restrictions imposed by neighboring cells and may lose positional information[60,61]. In contrast, cells in meristematic tissues are tightly connected by thin primary cell walls and intercellular filaments through which mobile transcription factors, signaling peptides, and phytohormones can regulate the proliferation and differentiation of surrounding cells[62−64]. A study of stem tip-induced SE in Arabidopsis thaliana reported the immediate expansion of the expression of several genes related to stem meristematic tissue in the stress response, which may provide cells with the ability to shift their fate from shoot to embryo (Supplementary Table S1)[65].
Medium composition and culture conditions
-
The culture medium plays a crucial role in the isolated regeneration culture of medicinal plants, providing the necessary nutrients but also influencing the regeneration efficiency of plants through the regulation of different components. In tissue culture, MS medium is widely used as a basic medium because of its high nutrient content and is particularly suitable for the growth and development of the shoots and roots of medicinal plants. However, different species or tissues may require different culture medium[2,66]. For example, a study reported that the proliferation coefficient and differentiation rate of Lonicera macranthoides cultured on MS medium were greater than those of L. macranthoides cultured on other medium[67]. In addition, a study reported that 1/2 MS medium was significantly better than MS medium at promoting the growth of tissue culture-generated seedlings (Supplementary Table S1)[27].
The carbon source is another key component of the culture medium that provides energy and regulates the osmotic environment for plant tissue culture. Commonly used carbon sources include glucose, sucrose, and maltose, which have important effects on the formation and maturation of somatic embryos[68]. Sucrose is considered one of the best carbon sources because of its ability to promote the growth and differentiation of explants[69]. In flax healing tissue culture, sucrose is used as a carbon source at an optimal concentration of 30 g/L (Supplementary Table S1)[70].
Light and temperature are also important factors affecting the regeneration of medicinal plants in vitro. Light plays an inducing role in the growth and differentiation of cells, tissues and organs, but excessive light or an improper photoperiod may be detrimental to the formation and maintenance of healing tissues. For example, the oxidation of phenolic compounds under light conditions may lead to tissue browning and affect somatic embryo formation[71]. A proper dark culture time is beneficial for increasing the callus volume of perilla frutescens. The temperature affects the respiratory rate and metabolic reactions, and the optimum temperature for medicinal plants is generally approximately 25 °C (Supplementary Table S1); however, different medicinal plants, such as Linum usitatissimum, Scutellaria baicalensis, Asarum heterotropoides, and P. grandiflorus, need different temperature conditions to promote callus growth and organ differentiation.
In the culture of medicinal plants in vitro, the physiological status and genetic differences of explants can be optimized for optimal regeneration by adjusting the composition of the culture medium. These methods include the selection of appropriate conditions, such as basic medium, carbon sources, photoperiods, and temperatures, to suit the specific needs of different medicinal plants (Supplementary Table S1).
-
Early studies on plant cell growth and development focused mainly on the role of plant hormones. However, since the end of the 20th century, with the development of molecular genetics and transcriptomic technologies, researchers have begun to pay more attention to transcription factors and the multiple signaling pathways and expression patterns they regulate. Recent studies at the molecular level have shown that plant cell dedifferentiation and morphogenesis depend on the orderly and correct expression of specific transcription factors[72]. These processes, including healing tissue formation, de novo shoot and root regeneration, and SE, are regulated by finely tuned hormonal and abiotic stress signals, in which cellular totipotent transcription factors play crucial roles[73].
Epigenetic modifications, including DNA methylation and histone modifications, also play important roles in healing tissue formation and organ regeneration, especially in the restoration of somatic cells to pluripotent healing tissue cells induced by plant hormones and injury[74]. In addition, plant regulatory ncRNAs are essential for healing tissue induction because they influence gene expression and protein translation and participate in transcriptional and posttranscriptional regulation[75].
Although studies on the molecular mechanisms of hormone regulation, epigenetic regulation and ncRNA regulation are rare in the field of medicinal plant regeneration, we can provide a theoretical basis and reference for future studies on the molecular mechanisms of medicinal plant regeneration by summarizing the regulatory mechanisms in other plants.
Molecular mechanisms of SE in plants
-
SE is a classical dedifferentiation process that allows differentiated somatic cells to revert to embryonic stem cells with totipotency, providing the basis for totipotency and regeneration in multicellular organisms. Recent studies have revealed that this process not only involves multiple transcription factors and hormonal signaling pathways but is also closely related to epigenetic regulation and the fine regulation of non-coding RNAs (ncRNAs) (Fig. 3).
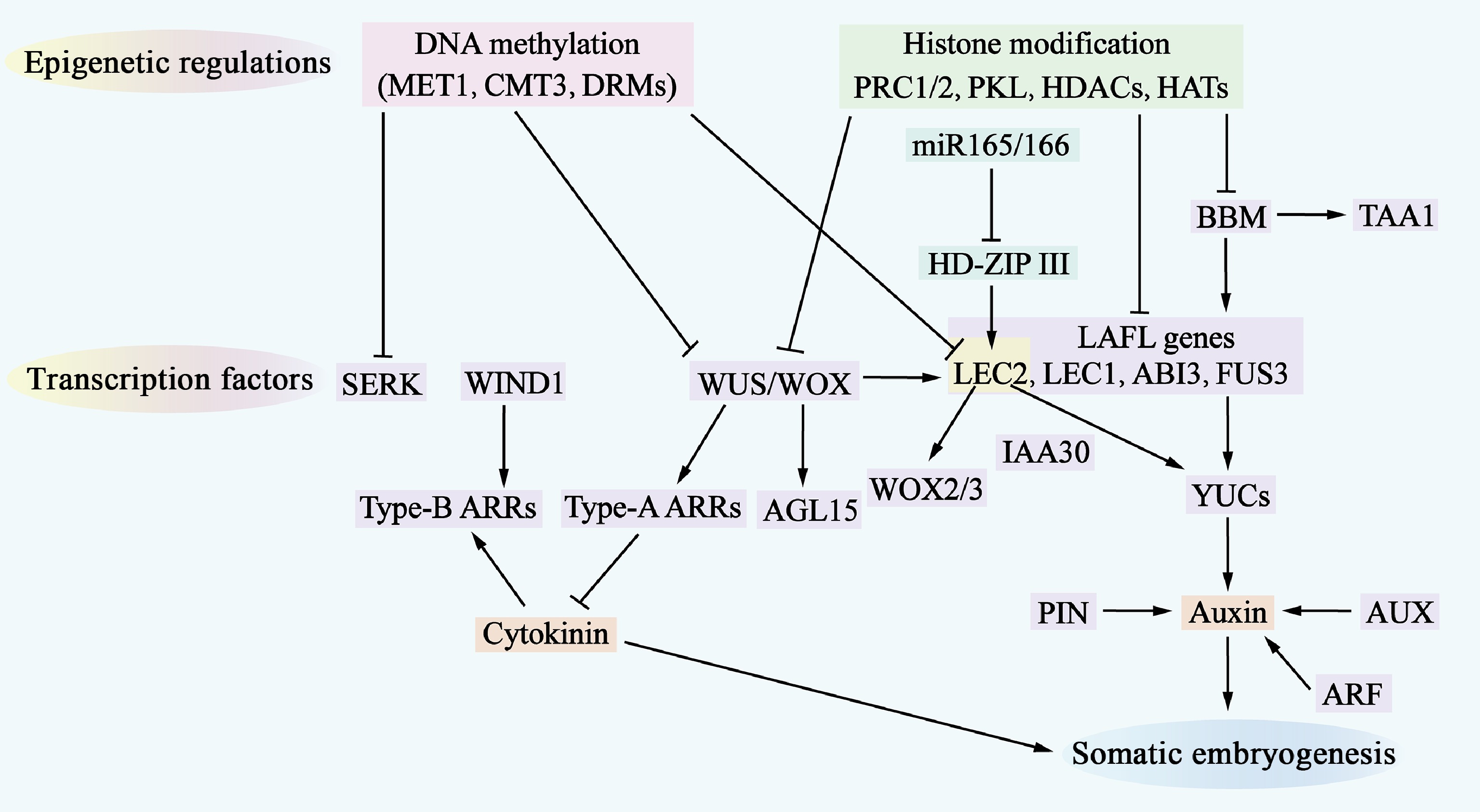
Figure 3.
Molecular mechanisms of somatic embryogenesis. The process of somatic embryogenesis is influenced by epigenetic regulation, transcription factors, and hormone signaling pathways. Epigenetic regulation, which includes DNA methylation (indicated by a pink shadow) and histone modifications (indicated by a green shadow), represses the access of transcription factors to gene-promoter regions, thereby inhibiting the expression of genes involved in somatic embryogenesis. Numerous transcription factors (indicated by a purple shadow) are involved in this regulatory network, where they also regulate each other and activate downstream auxin and cytokinin (CK) signaling pathways. Additionally, miR-165/-166 (indicated by a cyan shadow) are involved in regulating somatic embryogenesis.
In terms of transcription factors, Plethora (PLT), BABY BOOM (BBM), Leafy cotyledon 1 (LEC1), LEC2, RWP-RK RWP-RK DOMAIN-CONTAINING 4(RKD4)/GROUNDED (GRD), At-hook motif containing nuclear localized15 (AHL15), WUSCHEL (WUS), FUSCA 3 (FUS3), and Abscisic acid insensitive 3 (ABI3) have been identified as key factors regulating SE (Supplementary Table S3). These transcription factors are important regulators that drive cell fate transitions and can be considered representative of totipotency-associated transcription factors[36].
BBM and PLT, as members of the AP2/ERF family, play crucial roles in the growth and development of embryonic and root meristematic tissues in A. thaliana[76,77]. The ectopic expression of the BBM gene induced SE and regenerated seedlings without exogenous growth regulators in both A. thaliana and Brassica napus, highlighting the central regulatory role of BBM in the development of plant embryos[78]. In addition, this study further revealed the specific activation of the BBM gene during the transformation of plant somatic cells to embryonic cells and its precursor role in signaling pathways to promote cell differentiation and somatic embryo formation (Supplementary Table S3)[79].
Transcription factors such as ABI3, FUS3 and LEC2 encode proteins containing the plant-specific b3 structural domain and belong to the AFL subfamily[80,81]. These proteins, together with the LAFL complex formed by LEC1, are involved in the activation of cellular allosteric transcription factors. BBM is located upstream of these transcription factors and can activate cellular allosteric transcription factors, such as LEC1, LEC2, and agamous-like 15 (AGL15), indirectly promoting the expression of auxins signaling factors such as YUCCA (YUC) and the AUX/IAA factor IAA30. BBM directly binds to the promoter regions of the YUC and tryptophan aminotransferase of Arabidopsis 1 (TAA1) genes, driving the upregulation of their expression, increasing auxin synthesis and promoting SE (Supplementary Table S3)[82,83] (Fig. 3).
WUS homology box transcription factors also play key roles in the regulation of embryonic cell fate. In A. thaliana, WUS overexpression promotes somatic embryo production under hormone-free conditions and upregulates the expression of LEC1, LEC2, and AGL15 during SE[78]. WOX2 and WOX3, downstream targets of LEC2, are essential for somatic embryo formation. Moreover, WOX2/3 are essential for SE, but their overexpression is not sufficient to induce somatic embryo formation (Supplementary Table S3)[84] (Fig. 3).
Wound-induced differentiation 1 (WIND 1), another APETALA2/ERF family transcription factor, is not directly involved in somatic embryo formation but plays an important role in healing tissue induction. WIND1 is located upstream of LEC2 during regeneration and is involved in cytokinin-specific responses rather than auxin biosynthesis and signaling through different hormonal pathways[9,84]. In particular, WUS represses negative regulators [type-A Arabidopsis response regulator (ARR) genes] of the CK response, whereas WIND1 stimulates the expression of positive regulators (type-B ARR genes) of the CK response (Supplementary Table S3)[72,85] (Fig. 3).
Epigenetic regulation plays a key role in maintaining somatic cell identity and suppressing embryo-specific gene expression. DNA methylation and histone modification are important mechanisms that regulate gene expression and determine cell fate[86]. Mutation of methyltransferase 1 (MET1) affects the expression of the auxin efflux vector Pin-formed 1 (PIN1) and leads to abnormalities during SE[87]. The methylation levels of somatic embryogenesis receptor-like kinase (SERK), LEC2 and WUS in embryogenic healing tissues suggest a potential role for these genes in the regulation of SE[88]. In addition to DNA methylation, histone modifications, including methylation, acetylation and ubiquitination, also play important roles in regulating SE. A study reported that Polycomb repressive complex (PRC) 1 and PRC2 are required to establish and maintain stable epigenetic repression in response to developmental or environmental signals and that PRC1 and PRC2 repress the expression of embryo-specific genes, including LAFL, AGL15, WUSCHEL-RELATED HOMEOBOX 5 (WOX5), BBM, and PIN1[89]. In addition, PICKLE (PKL) plays an important role in preventing the generation of embryonic traits in somatic cells and is an epigenetic factor that plays a key role similar to that of PRC1 and PRC2[90]. A study further reported that PKL represses the expression of embryonic genes, including the LAFL genes, by promoting alterations in Histone 3 lysine 27 trimethylation (H3K27me3)[91]. In addition, histone acetylation regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs) plays a key role in SE. The HDAC inhibitor tricosin A upregulates the expression of genes related to embryogenesis, including LEC1, FUS3 and ABI3 (Supplementary Table S3)[92] (Fig. 3).
ncRNAs, including miRNAs and long-chain non-coding RNAs, also play a role in somatic cell embryogenesis. Specific miRNAs are involved in the regulation of SE through the modulation of phytohormone signaling pathways[74]. A study revealed that the regulation of RNA metabolism is essential for in vitro phytocellular dedifferentiation and that high levels of mini-nucleolar RNAs are required for in vitro cellular dedifferentiation and organogenesis in A. thaliana[93].
Molecular regulatory mechanisms of phytohormone-induced callus formation
-
The formation of pluripotent healing tissues is a complex process that usually begins with the division of column sheath cells in the xylem pole; this process is similar to lateral root formation and involves the same molecular mechanisms[94] (Fig. 4). Auxin is a key hormone involved in the regulation of healing tissue formation and lateral root formation[95], which prompted us to hypothesize that some components of auxin signaling or downstream factors involved in lateral root formation may also play important roles in healing tissue regulation.
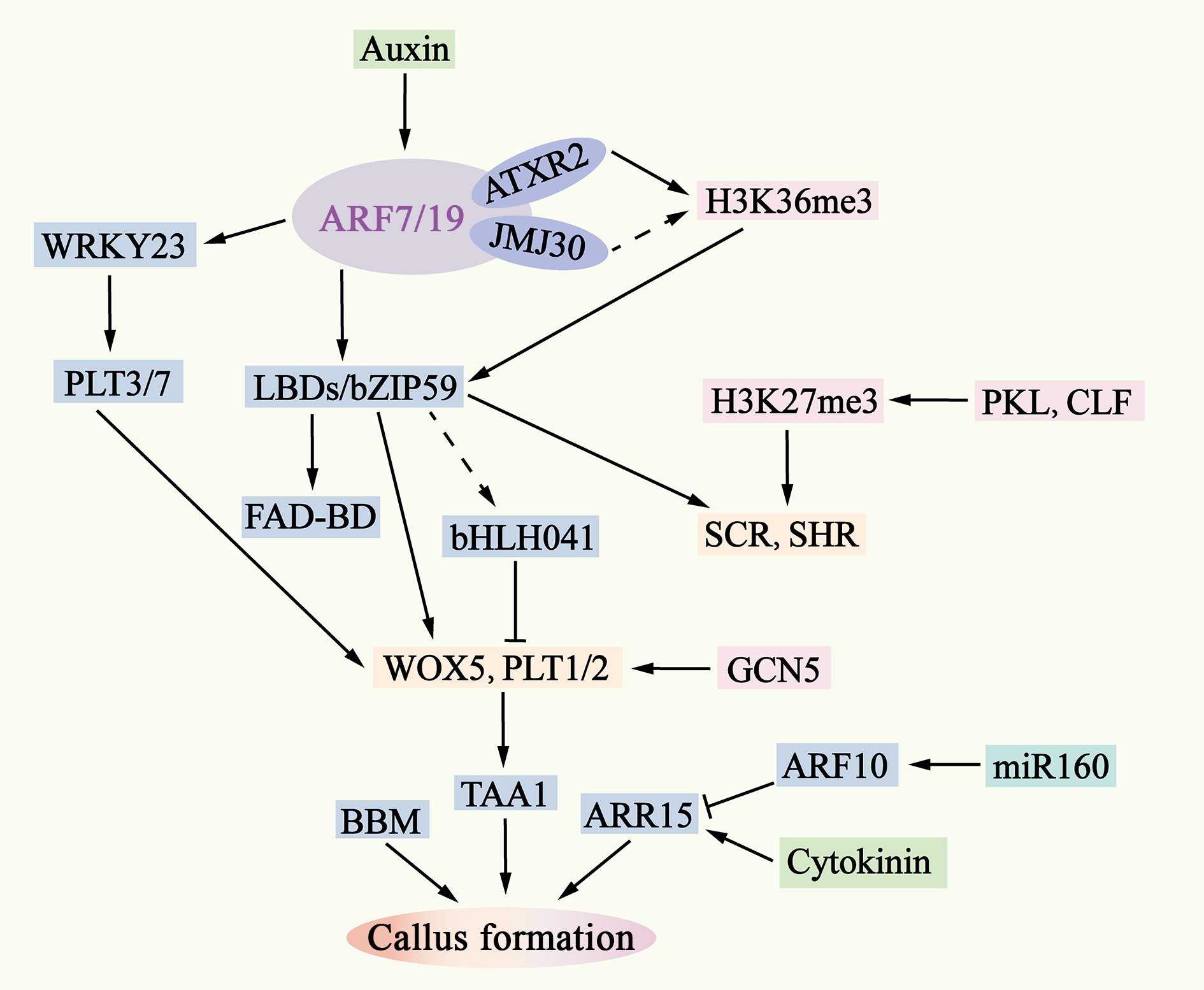
Figure 4.
Molecular mechanisms of pluripotent callus formation. The ability of auxin to activate ARF7/19 is also regulated by epigenetic factors such as AXTR2 and JMJ30. Downstream of ARF7/19, LBD-bZIP59 and WRKY23 activate the expression of genes such as WOX5, PLT1/2, SHR, and SCR by removing bHLH041 and PLT3/7. The interaction between WOX5 and PLT1/2 enhances the expression of TAA1, leading to an increase in endogenous auxin levels in the callus and inducing the formation of pluripotent calli. The removal is indicated by dashed lines.
In A. thaliana, auxin response factor (ARF)7 and ARF19, in conjunction with SOLITARY ROOT (SLR/IAA14), were initially identified as important regulators of lateral root formation, and they are also required for auxin-induced healing tissue formation[96]. When the function of ARF7/19 or SLR is disrupted or mutated, healing tissue formation is significantly inhibited[97]. In addition, the LOB-DOMAIN transcription factor lateral organ boundaries domain (LBD)16, LBD17, LBD18 and LBD29, which are downstream targets of the ARF7/19-IAA14 module, are likewise essential for healing tissue formation. The transcription factor bZIP59 synergizes with these LBD proteins, and growth factors promote cell fate transition by stabilizing bZIP59 and enhancing its interaction with LBDs[97]. Further studies revealed that LBDs form a heterodimeric complex with bZIP59, which directly activates, among other enzymes, FAD-BINDING BERBERINE (FAD-BD), which encodes a BBE-like enzyme involved in cell wall metabolism and is a direct target of LBD16 to promote lateral root emergence[98,99]. In transgenic seedlings overexpressing LBD or bZIP59, ectopic activation of the root meristem tissue regulators WOX5, PLT1, PLT2, SCARECROW (SCR) and SHORTROOT (SHR) (Fig. 4), which are essential for the maintenance of healing tissue pluripotency and subsequent neoplastic regeneration, was observed (Supplementary
Table S3)[97].The physical interaction between WOX5 and PLT1/2 promotes the expression of the auxin biosynthesis gene TAA1, which is essential for maintaining the pluripotency of healing tissue cells[100]. In addition, the loss of function of the PLT3/5/7 genes does not affect the formation of healing tissues but hampers the ability of healing tissues to form preemergent cells in a subsequent regeneration program[101]. A. thaliana WRKY23 and bHLH041 act as transcriptional activators and repressors downstream of ARF7/19 and are responsible for the activation of root stem cytokines, which establish healing tissue pluripotency. WRKY23 directly targets and activates the transcription of PLT3/7, whereas the LBD-induced removal of bHLH041 represses the transcription of PLT1/2 and WOX5[102] (Fig. 4 & Supplementary Table S3).
In addition to the above genes, the overexpression of BBM in different plant species also promotes cell proliferation and regeneration. For example, the overexpression of A. thaliana BBM in Nicotiana tabacum induces callus formation; the overexpression of GmBBM7 in soybean promotes the growth of calli and roots; and the overexpression of AtBBM in poplar leads to the formation of somatic embryos[73].
The in vitro of healing tissue involves cell fate transitions and reprogramming at the genome-wide level. Epigenetic regulation plays an important role in auxin-induced healing of tissue. For example, Arabidopsis Trithoprax-related 2 (ATXR2) and JMJ30 activate LBD gene expression by regulating histone modifications, whereas the histone acetyltransferase GCN5 acquires pluripotency by catalyzing histone acetylation at the root meristem gene locus[103]. In addition, chromatin remodeling factors such as PKL and CURLY LEAF (CLF) are involved in the establishment of healing tissue pluripotency by controlling the expression of root stem cell regulators[64] (Fig. 4 & Supplementary Table S3).
ncRNAs, such as miR160, also play key roles in healing tissue induction and plant cell dedifferentiation. miR160 and its target gene ARF10 are critical in the process of healing tissue formation in vitro (Fig. 4 & Supplementary Table S3). More prolific and faster healing tissue formation was observed in specimens with miR160-resistant forms of ARF10 (mARF10), whereas cell lines overexpressing miR160 had slower and fewer healing tissues[104].
Molecular mechanisms of phytohormone-induced root regeneration
-
In tissue culture, the transfer of healing tissues to a root-induction medium containing relatively high concentrations of auxin induces the development of new rooting organs. The inhibition of polar auxin transport blocks the rooting process, which highlights the key role of auxin in regulating the development of new root organs[105]. In addition, the YUC gene family (YUC1, YUC11, YUC8, and YUC9), which is involved in the biosynthesis of auxins, has been shown to inhibit the expression of WOX11 in receptor cells (Supplementary Table S3)[106].
Recent transcriptomic, epigenomic and cell lineage analyses of healing tissues revealed similar genetic pathways for healing tissue formation and neoplastic root organogenesis. The de novo process of root organogenesis can be divided into two major steps: first, the transition from energetic to root-establishing cells, a process in which the expression of WOX11 is a hallmark event; and second, the transition from root-establishing to root-primary cells, marked by the expression of WOX5[107]. In the first step, auxin directly activates the expression of WOX11 and its homolog WOX12. WOX11/12 subsequently further promotes the expression of WOX5 and LBD16, and then LBD16 is responsible for activating the expression of WOX5, PLT1 and PLT2[108] (Fig. 5 & Supplementary Table S3).
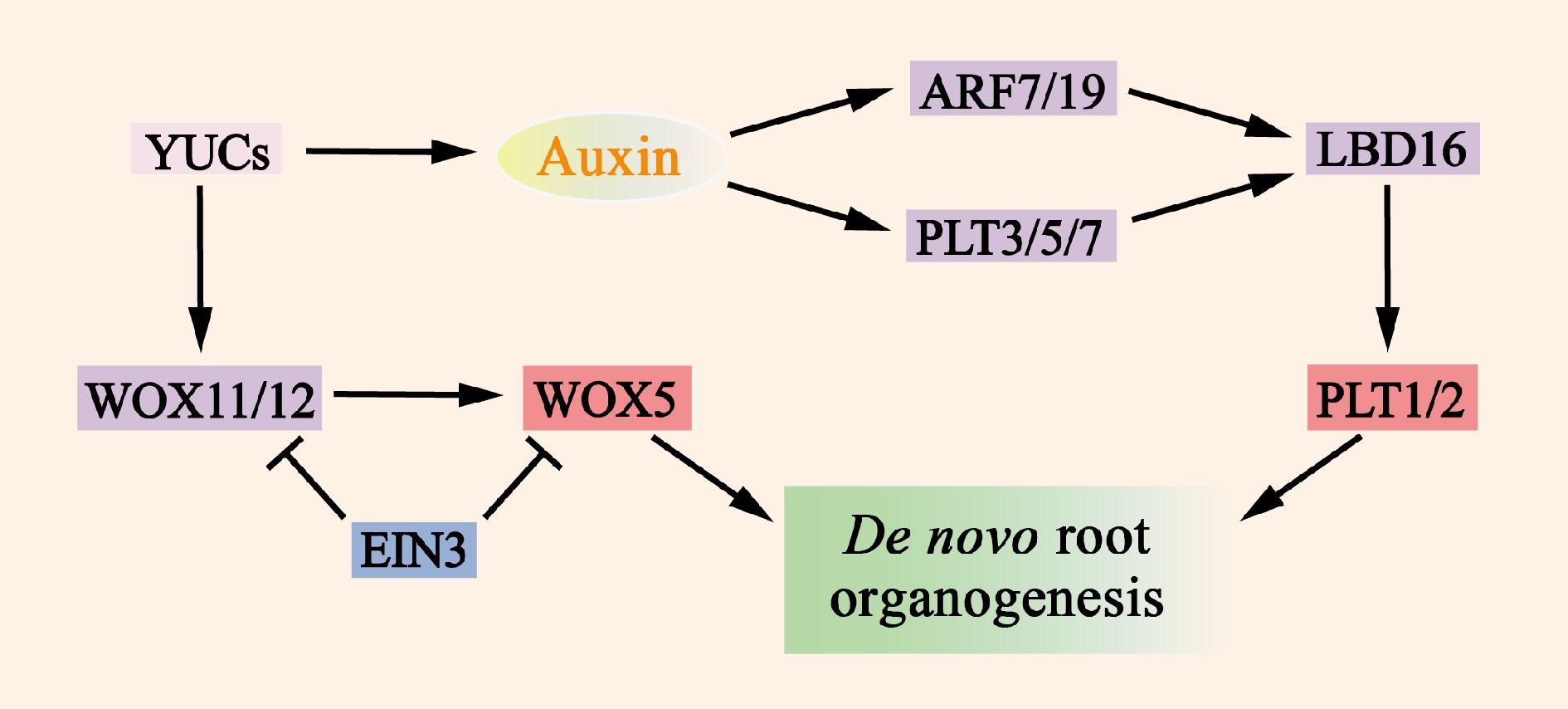
Figure 5.
Molecular mechanisms of de novo root organogenesis. Auxin, which is mediated by YUC, serves as a key regulatory factor that activates the expression of WOX11/12, ARF7/19, and PLT3/5/7. The translation products of these genes then directly or indirectly promote the expression of WOX5 and PLT1/2, which in turn induce the formation of de novo root organogenesis. The transcription factor EIN3 significantly reduces the frequency of new root organs by inhibiting the transcription of WOX11 and WOX5.
The transcription factor EIN3 significantly reduces the frequency of newly rooted organs by repressing the transcription of WOX11 and WOX5. This finding is consistent with the observations that the activity of EIN3 increases with the age of the explant and that younger organs have greater regenerative capacity[109]. As mentioned previously, growth factors also induce the expression of PLT3, PLT5, and PLT7 and regulate the expression of downstream root meristem organization marker genes. In addition to the WOX11/12 and PLT genes, the auxin response factors ARF7 and ARF19 can target and activate the expression of LBD16 (Fig. 5 & Supplementary Table S3), which promotes expression during root regeneration.
Although studies on the molecular mechanisms of epigenetic modifications and ncRNAs in the induction of de novo root regeneration are still relatively limited, further exploration in these areas will undoubtedly provide a deeper understanding. With further research, the elucidation of these regulatory mechanisms will help optimize plant tissue culture conditions, improve regeneration efficiency, and provide new strategies for plant biotechnology applications.
Molecular mechanisms of phytohormone-induced shoot regeneration
-
Cultivation of healing tissues in a cytokinin-rich medium induces continued cell division and proliferation mediated by cytokinin to form cell populations and promote subsequent differentiation, which marks the establishment of the stem cell ecological niche[110]. The maintenance of stem homeostasis is achieved through two main regulatory pathways, WUS–Clavata 3 (CLV3) and Shoot meristemless (STM)–cup-shaped cotyledon (CUC) (Fig. 6 & Supplementary Table S3), which are decisive factors in the early stage of stem cell ecological niche construction. The WUS gene begins to be expressed 2 to 3 d after tissue culture[111], and its initial expression is a hallmark of the establishment of shoot progenitor cells, which is the most critical molecular event in the process of de novo shoot organogenesis. WUS mutants completely lose their regenerative capacity, whereas WUS overexpression results in the ectopic formation of shoots, confirming the necessity of WUS in the regeneration of nascent shoots[112].
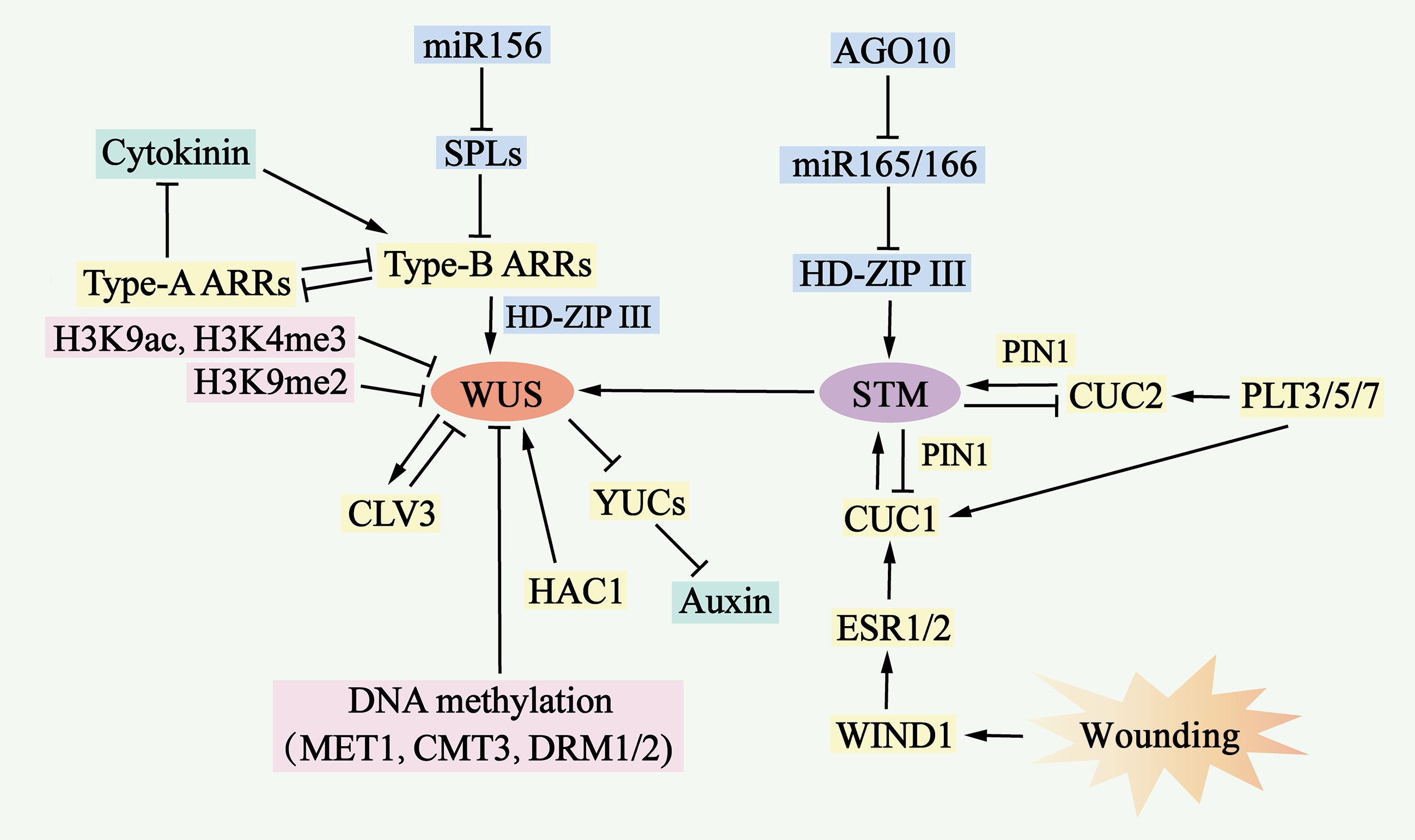
Figure 6.
Molecular mechanisms of de novo shoot organogenesis. During the process of de novo shoot organogenesis, two pathways, the WUS-CLV3 pathway and the STM-CUC pathway, establish negative feedback loops and play critical regulatory roles. The WUS-CLV3 pathway is regulated primarily by DNA methylation, histone modification, and hormone signaling. Cytokinin (CK) activates the expression of type B ARRs, which in turn stimulates WUS expression, whereas type B ARRs repress YUC-mediated auxin biosynthesis. In the STM-CUC pathway, STM expression is promoted by CUC1 and CUC2, both of which are upregulated by PLT3/5/7, ESR1, ESR2, WIND1, and PIN1. Moreover, WUS and STM interact directly to activate CLV3 expression, suggesting that the two pathways converge and coordinate to control shoot regeneration.
WUS promotes the expression of the encoded signaling peptide CLV3, which represses WUS expression through a negative feedback loop, serving as a negative feedback mechanism that plays a key role in maintaining stem cell populations. Similarly, the STM gene is expressed in stem meristematic tissues and represses the expression of CUC1 and CUC2, which in turn activates STM expression to maintain stem meristematic tissues[113]. The WUS–CLV3 pathway is regulated by DNA methylation, histone modification, and hormone signaling (Fig. 6 & Supplementary Table S3).
The auxin and cytokinin signaling pathways jointly influence WUS expression. B-type response regulators (ARR1, ARR2, ARR10 and ARR12), as transcriptional activators of cytokinin signaling, activate the expression of WUS by directly binding to its promoter and inhibiting YUC gene-mediated auxin accumulation, which further promotes WUS expression[111]. In contrast, a-type response regulators (ARR5, ARR6, ARR7 and ARR15), negative regulators of cytokinin signaling, are regulated by b-type response regulators, which form a negative feedback loop to inhibit shoot regeneration by interfering with the function of b-type response regulators[114] (Fig. 6 & Supplementary Table S3). In addition, type B response regulators interact with the HD-ZIP III protein to form a transcription complex, which specifically activates the expression of WUS[110].
In the STM–CUC pathway, a negative feedback loop between STM and CUC plays a key role in regulating neoplastic stem organogenesis. CUC proteins are essential for the establishment of the shoot progenitor system[77]. The CUC-induced polar localization of PIN1 determines shoot progenitor location, and the increased polarity of PIN promotes STM expression in the progenitor system[112]. In addition, PLT3, PLT5, and PLT7 upregulate the expression of CUC1 and CUC2 during shoot regeneration. These PLT proteins control shoot regeneration through a two-step mechanism: first, they activate the expression of PLT1 and PLT2 during pluripotent guaiac tissue formation to increase competence; second, they accomplish regeneration through the regulation of CUC[115]. Enhancer of shoot regeneration (ESR)1 and ESR2 act as upstream regulators of CUC genes during neoplastic stem organogenesis by directly binding to their promoters to activate expression. In addition, ESR1 expression is regulated by WIND1, linking wound signaling to shoot regeneration[116] (Fig. 6 & Supplementary Table S3).
Both the WUS–CLV3 and STM–CUC pathways are required for stem cell development during de novo shoot organogenesis. Recent studies have reported that these two pathways are coordinated through direct interactions between the WUS and STM proteins. STM directly activates CLV3 expression by binding to the promoter at a site different from WUS. WUS–STM interactions enhance WUS binding to the CLV3 promoter and CLV3 transcriptional activation, suggesting that CLV3 is simultaneously regulated by WUS, STM, and WUS–STM complexes[117] (Fig. 6 & Supplementary Table S3).
Epigenetic modifications can regulate gene transcription. The WUS locus is a site of DNA methylation and inhibitory histone modifications such as H3K27me3[111]. Under wild-type conditions, the WUS promoter is highly methylated; however, mutations in MET1, Chromomethylase 3 (CMT3), Domain rearranged methyltransferase 1 (DRM1) and DRM2 result in the deletion or reduction in DNA methylation in the regulatory region of the WUS promoter, which increases the expression of WUS and the shoot regeneration rate[114]. Neonatal shoot regeneration involves different histone modification sites in WUS, and the abundance of histone 3 lysine 9 acetylation (H3K9ac) and histone 3 lysine 4 trimethylation (H3K4me3) at the WUS locus increases during stem regeneration, whereas the abundance of the H3K9me2 repressor at the WUS locus decreases during stem regeneration[118]. HAC1 and Lysine-specific demethylase 1-like 3 activate WUS transcription and increase shoot yield[119]. The removal of H3K27me3 from the WUS locus appears to be cytokinesis dependent, and olomoucine delays the decrease in H3K27me3 levels at the WUS locus and induces WUS expression[111] (Fig. 6 & Supplementary Table S3).
In addition, miR-156 targets SPL mRNA and decreases b-type ARR activity in an age-dependent manner[120]. In young explants, the expression level of miR156 inhibit SPL expression level, which increases b-type ARR activity and shoot regeneration capacity. Moreover, miR-165/-166 inhibits stem regeneration by splicing and decreasing the translation of the mRNA encoding the HD-ZIP III protein[121]. The Argonaute 10 (AGO 10) gene inhibits stem regeneration by repressing miR-165/166 activity (Fig. 6 & Supplementary Table S3).
-
Medicinal plants are important natural resources for humans; they can not only cure diseases but also enhance human immunity and prevent the occurrence of diseases. With the frequent occurrence of global public health crises and increasing attention given to life and health, TCM has ushered in a critical period of revitalization and development. Medicinal plant resources constitute the cornerstone for promoting the high-quality development of the TCM industry. According to preliminary statistics from the Fourth National Census of Chinese Medicine Resources, there are more than 13,000 kinds of medicinal plants in China, of which more than 200 commonly used bulk Chinese herbal medicine are cultivated on a large scale[122]. Strengthening the breeding of good seeds of local herbs and improving the quality of Chinese herbal medicines from the source are key to ensuring the safety, efficacy and stability of clinical medication. Biological breeding, especially the application of gene editing technology, will lead to changes in the molecular breeding technology utilized for TCM. However, research on the regeneration of medicinal plants has focused mainly on the application level; additionally, there is a lack of systematic and in-depth exploration of regeneration mechanisms and future development, and several challenges have limited the widespread use of medicinal plant regeneration.
First, improving the quality of Chinese herbal medicines to ensure their clinical efficacy is a core issue of TCM resources. The development of the Chinese medicinal seed industry is key to ensuring the yield and quality of Chinese herbal medicine. The breeding of Chinese herbal medicine is at the stage of original domestication selection, hybridization and molecular breeding, and the molecular design of TCM is still in its infancy[123]. Recently, new varieties of Chinese herbal medicine have been cultivated, such as P. ginseng 'Xinkaihe No. 1' and S. miltiorrhiza 'chuan Danshen No. 1', all of which are obtained via conventional methods such as systematic selection and cross-breeding, and their breeding efficiency is low[124]. One of the main reasons for the relative lag in the development of the molecular breeding of medicinal plants compared with that of common crops is that medicinal plants generally suffer from poor regeneration and low genetic transformation efficiency, characteristics that are strongly influenced by species and genotypes[125].
Second, explant browning is a major factor that hinders the growth and differentiation of medicinal plants. Browning refers to the activation of polyphenol oxidase in the explant culture process, which oxidizes phenolic substances into quinones, leading to browning of the culture medium and hindering the growth and differentiation of medicinal plants[126]. Different plant genotypes, sampling sites and physiological states are the main factors leading to browning. Therefore, preventing the browning of explants to ensure their normal growth and differentiation during the regeneration of cultured medicinal plants is crucial.
Finally, the tissue culture systems themselves have several limitations. Tissue culture technology relies on the precise control of the concentration ratio of auxin and cytokinins, which play important roles in genetic transformation[55,125]. However, this process requires strict maintenance under aseptic conditions, growth medium with specific carbon sources and hormone ratios, and controlled environmental conditions with appropriate light and temperature. These conditions are demanding and require abundant resources and expertise, making the tissue culture process labor intensive and costly[127]. In addition, methods that rely on tissue culture are time-consuming and carry the risk of somatic cell asexual lineage variation, and different plant species and genotypes respond differently to tissue culture conditions, leading to wide variations in regeneration efficiency[19,128].
Future directions for enhancing the efficiency of medicinal plant regeneration
-
Enhancing the regenerative capacity of different species of medicinal plants requires a multifaceted approach. Key strategies include optimizing tissue culture systems, incorporating morphogenetic factors, exploring new ways to bypass traditional tissue culture, and using gene editing technology to improve plant cell regeneration efficiency.
First, optimizing tissue culture systems is the basis for improving the regeneration ability of medicinal plants. Various factors in the tissue culture environment, such as the basic medium, carbon source, hormone concentration and environmental conditions (light intensity and photoperiod), profoundly affect regeneration efficiency. For example, MS medium is suitable for the proliferation and growth of A. paniculata shoots, with a proliferation rate of 83.3% and good growth[129]. However, five different basic medium (MS, 1/2 MS, MT, H and B5) were compared and optimized, and it was reported that 1/2 MS was better for the growth of A. paniculata seedlings. Finally, the effects of MS, N6 and Nistch medium on anther culture were compared and optimized, and it is reported that N6 medium was the most suitable for the culture of A. paniculata anthers.
Morphogenetic factors are powerful catalysts for increasing the rate of regeneration by influencing key cellular reprogramming and differentiation pathways. Transcription factors such as WUS2 and BBM have a remarkable ability to stimulate somatic cells, inducing them to form embryos that subsequently develop into full plants. Furthermore, some transcription factors have been shown to promote plant regeneration when combined with their cofactors. For example, the fusion of Triticum aestivum GROWTH-REGULATING FACTOR 4 (GRF4) and its auxiliary factor GRF-INTERACTING FACTOR 1 (GIF1) has been shown to be very effective in increasing the regeneration speed and efficiency of T. aestivum, Secale cereale and Oryza sativa, transgenic GRF4-GIF1 plants were fertile without obvious defects. In addition, GRF4-GIF1 enhanced wheat regeneration in the absence of exogenous cytokinin, facilitating transgenic selection in the absence of selection markers[130]. The combination of these morphogenetic regulatory factors is particularly beneficial to plant species that are difficult to regenerate or have a long regeneration cycle.
New methods that bypass traditional tissue culture have attracted significant attention because they have the potential to revolutionize crop regeneration. Techniques such as the flower dip method and the cut-and-dip budburst (CDB) system offer species-specific alternatives to transform seeds and direct root transformation. CDB delivery systems use Agrobacterium rhizogenes to inoculate explants, generating transformed roots that produce transformed shoots. A variety of plant species in multiple plant families have been successfully transformed through CBD, including two herbaceous plants (Taraxacum kok-saghyz and Coronalla varia), a tuberous root plant (Ipomoea batatas), three woody plants (Ailanthus altissima, Aralia elata, and Clerodendrum chinense), and three succulents (Kalanche blossfeldiana, Crassula arborescens, and Sansevieria trifasciata). The CDB method allowed efficient transformation or gene editing in these plants using a very simple explant dipping protocol, under non-sterile conditions, without the need for tissue culture. In addition, large numbers of plants might be able to be genetically modified using CDB. These methods simplify the genetic modification process and reduce time and resource consumption[131,132]. In addition, innovative gene editing strategies, such as the direct injection of WUS2 and IPT into plants along with gene editing reagents, have opened new insights for tissue culture-independent gene editing[133]. Viral vectors such as TRV and Barley Stripe Mosaic Virus (BSMV) enable heritable and DNA-free gene editing by efficiently delivering gene editing materials into germ cells. In planta particle bombardment (iPB) and nanoparticle technologies also have the potential to simplify transformation[134].
Gene editing technologies have greatly improved the genetic improvement and regeneration efficiency of medicinal plants. CRISPR/Cas9 and other gene editing technologies improve plant cell regeneration[135]. Regeneration efficiency can be improved by gene editing, which promotes dedifferentiation and redifferentiation of plant cells. The CRISPR/Cas9 system allows precise gene modification, including single base substitution, and can be used to control the regeneration of medicinal plants and cultivate species with specific medicinal properties. The use of high-fidelity Cas9 mutants or optimized sgRNA designs will reduce off-target effects. The TnpB and IscB technologies, for example, offer a low risk of off-targeting, improving gene editing safety and specificity. Regeneration of medicinal plants with this method also reduces the need to modify non-target genes[136,137]. Gene editing has a wide range of potential applications in the regeneration of medicinal plants. In the future, these technologies will play a greater role in medicinal plant regeneration due to their continuous improvements and optimizations.
These multifaceted strategies will expand the scope of crop regeneration, making them more accessible, efficient, and adaptable to different species and genotypes. Medicinal plant regeneration plays a key role in agricultural biotechnology, especially in combination with precision genome editing and synthetic biology concepts[11], and several promising directions have emerged that have the potential to overcome current challenges and revolutionize the field. By integrating genomics and high-throughput sequencing technologies, a deeper understanding of the genetic basis of regeneration efficiency is possible[138]. Pyramiding the multiple factors involved in regeneration can reveal new regulatory centers and interactions[139]. Exploring non-tissue culture methods can bypass species and genotype-dependent tissue culture processes for plant regeneration[131−133]. Through the exploration of these future directions, regeneration research and the application of medicinal plants will contribute to the breeding of high-quality varieties of Chinese herbal medicine and the stock of germplasm resources, promote the establishment of regeneration systems for rare and endangered medicinal plants, provide new ideas for the screening of novel regeneration-promoting drug molecules[140,141], foster the intelligent production of Chinese herbal medicine, promote high-quality integration of the discipline of Chinese herbal medicine with modern science, and further promote the preservation and innovation of TCM.
-
In this work, two regeneration pathways of medicinal plants during SE and tissue culture, as well as the environmental factors and molecular mechanisms affecting these pathways, were comprehensively discussed. The information discussed provides valuable references for scientific research and technological development in this field. Although some progress has been made, the regulatory mechanisms of plant regeneration still need to be studied in depth. In vitro, the regeneration of plants is a complex process, and our understanding of this process is still relatively limited; more comprehensive and in-depth studies are needed to reveal the full picture.
Although the regulatory networks involved in plant regeneration have been initially identified, there is a relative lack of research on these networks in medicinal plants, and how the players and signaling molecules within medicinal plants synergize to facilitate the various stages of regeneration is still unclear. Therefore, future studies should further explore regeneration mechanisms in medicinal plants.
In tissue culture, traditional methods to improve the efficiency of plant regeneration rely heavily on altering external environmental factors. Future research should combine the understanding of molecular mechanisms with these traditional methods to optimize plant regeneration. Notably, this paper outlines regeneration control factors in nonmedicinal plants; however, whether medicinal plants have the same molecular mechanisms remains to be verified.
Currently, rapid micropropagation and genetic transformation of many important crops and medicinal plants are still challenging. Therefore, the future direction of medicinal plant regeneration research may lie in the application of theoretical concepts of plant regeneration to agricultural practices to establish efficient regeneration systems and promote the development and industrialization of agricultural biotechnology.
This work was supported by the National Natural Science Foundation of China (82460743, 32170402), the Major Special Science and Technology Project of Yunnan Province (202304BI090009; 202403AK140082), Yunnan Characteristic Plant Extraction Laboratory (2022YKZY001), Yunnan Province Youth Talent Support Program (XDYC-QNRC-2022-0219), Program of Shanghai Academic/Technology Research Leader (23XD1423500), Organizational Key Research and Development Program of Shanghai University of Traditional Chinese Medicine (2023YZZ01).
-
The authors confirm contribution to the paper as follows: study conception and design: Zhao Y, Wang J; literatures collection and analysis: Wang J, Liang YL, Liu GZ; writing-original draft preparation: Wang J, Li CH, Zhao Y; writing-review & editing: Yang SC, Xiao Y, Zhao Y. picture preparation and drawing: Zhou PH, Li CH. All authors approved the final version of the manuscript.
-
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
-
The authors declare that they have no conflict of interest. Dr. Ying Xiao is the Editorial Board member of Medicinal Plant Biology who was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of Dr. Xiao and the research group.
-
# Authors contributed equally: Juan Wang, Pin-Han Zhou
- Supplementary Table S1 Factors Affecting Medicinal Plants Regeneration.
- Supplementary Table S2 Plant hormones regulate the regeneration system of medicinal plants.
- Supplementary Table S3 Genes involved in regeneration.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Wang J, Zhou PH, Li CH, Liang YL, Liu GZ, et al. 2024. Progress on medicinal plant regeneration and the road ahead. Medicinal Plant Biology 3: e030 doi: 10.48130/mpb-0024-0026
Progress on medicinal plant regeneration and the road ahead
- Received: 19 September 2024
- Revised: 14 October 2024
- Accepted: 21 October 2024
- Published online: 26 December 2024
Abstract: Medicinal plants, which are valuable to human beings, play indispensable roles in various fields, such as health care, health promotion, and quality of life enhancement. They are not only pillars of traditional medicine but also valuable sources of modern pharmaceutical research and innovation. Although China has a rich variety of medicinal plants, in recent years, the drastic reduction in wild medicinal plant resources due to over-exploitation and over-utilization has affected the quality of Chinese herbal medicine. Therefore, the development of efficient in vitro regeneration culture technology for medicinal plants is particularly urgent. Here, the main regeneration pathways of medicinal plants are discussed, scientific progress of medicinal plant regeneration culture reviewed, and the main factors affecting the regeneration of medicinal plants analyzed, including the molecular mechanism of phytohormones in inducing the regeneration process, as well as the challenges faced by medicinal plant regeneration technology and directions for future development. Moreover, the challenges and future directions of medicinal plant regeneration technology are summarized, allowing us to find new ideas for the establishment of regeneration systems for rare and endangered medicinal plants, the screening of new regeneration-promoting drug molecules, and the preservation of traditional Chinese medicine (TCM) and its innovation.













