-
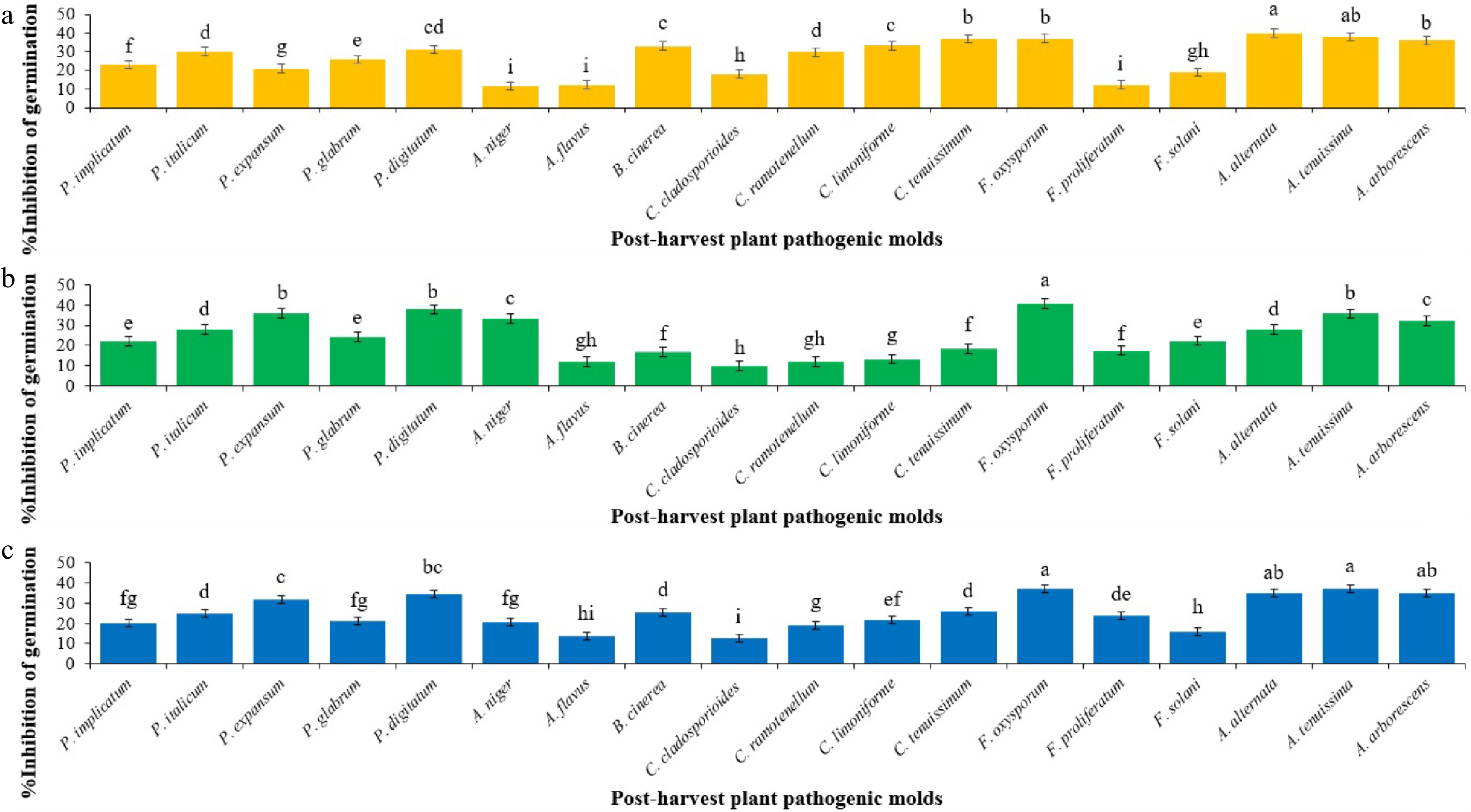
Figure 1.
Mean comparisons of the inhibition percentage of spore germination of post-harvest fungi as affected by secondary metabolites from initial spore germination stage of each Trichoderma sp. after 12 h. Values are means of five replicates. (a) The effect of T. atroviride on the inhibition of spore germination (Sum of squares: 7,659.592; df: 17; Mean square: 450.564; F-value: 169.764; p-value: 0.0001). (b) The effect of T. harzianum on the inhibition of spore germination (Sum of squares: 8316.157; df: 17; Mean square: 489.186; F-value: 142.419; p-value: 0.0001). (c) The effect of T. virens on the inhibition of spore germination (Sum of squares: 5410.919; df: 17; Mean square: 318.289; F-value: 86.543; p-value: 0.0001). Bars (standard error) with different letters indicate significant differences (Duncan range test subset for α = 0.05).
-
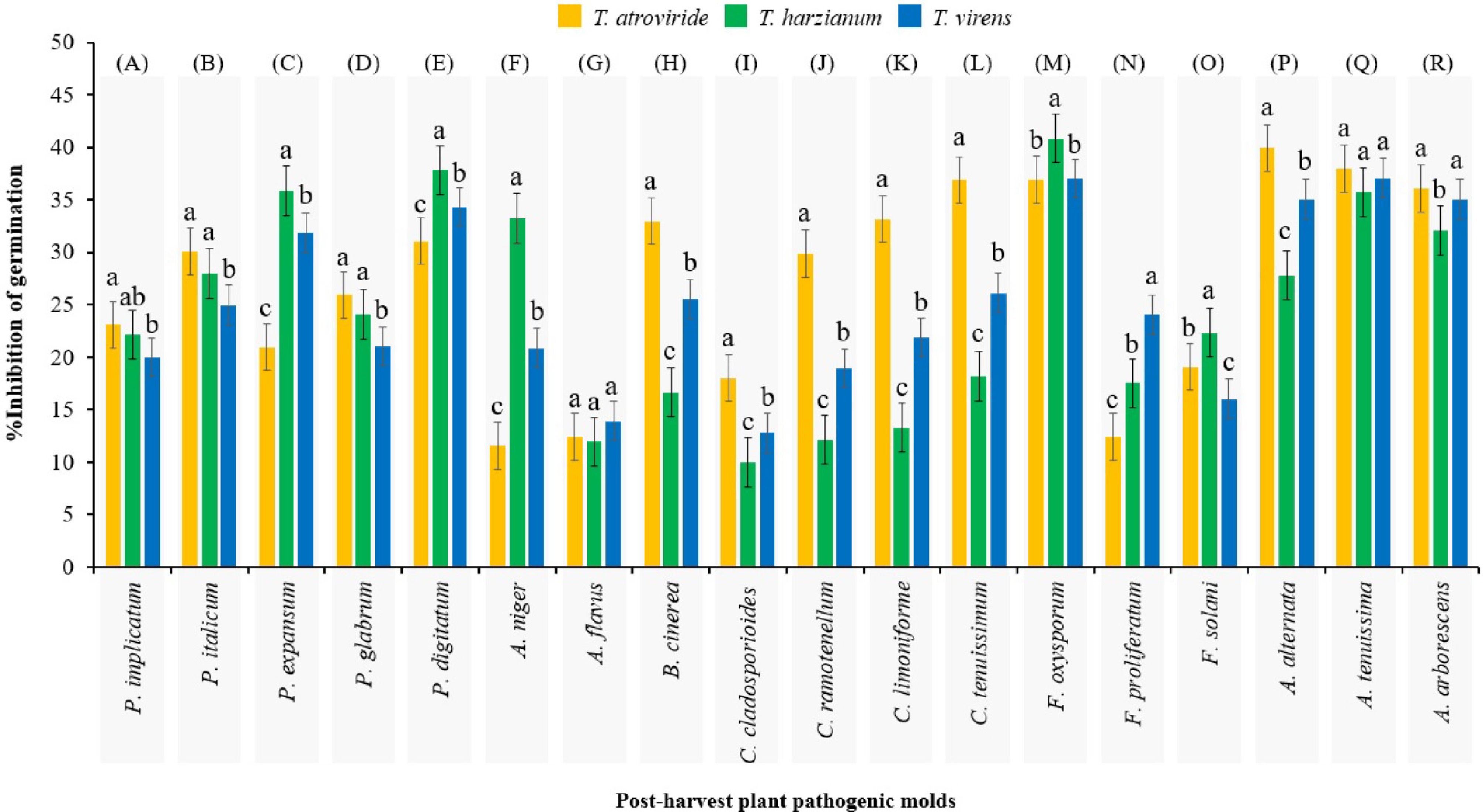
Figure 2.
Mean comparisons of the inhibition percentage of spore germination of each post-harvest fungus as affected by secondary metabolites from initial spore germination stage of Trichoderma spp. after 12 h. Values are means of five replicates. Bars (standard error) with different letters indicate significant differences. Sections (A)−(R) are independent experiments that grouped separately by Duncan's range test (subset for α = 0.05), which the results of their analysis of variance are presented in Table 2.
-
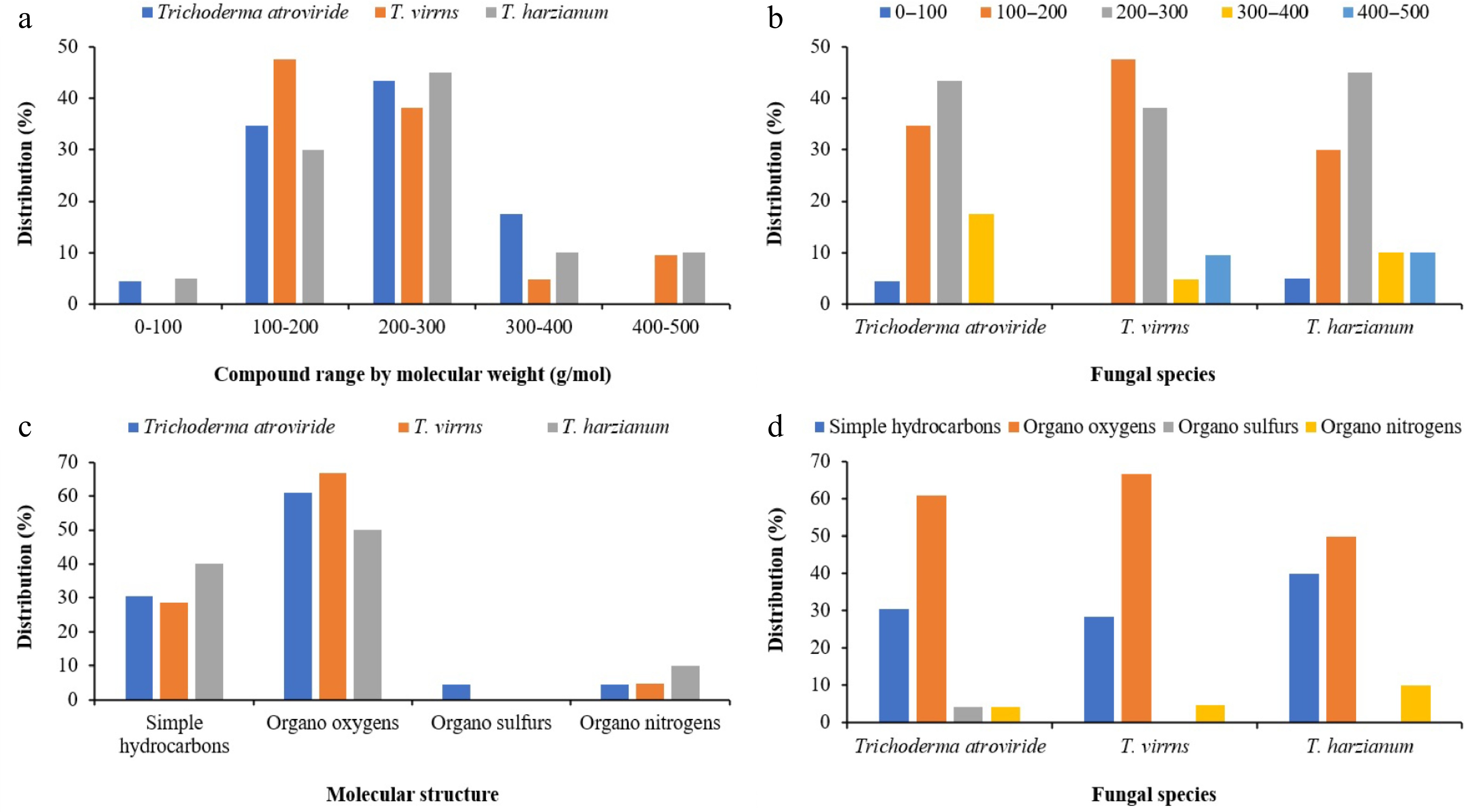
Figure 3.
Clustered column charts illustrating the distribution percentages of various metabolites across Trichoderma spp. extracts based on GC-MS analysis. (a) and (b) represent the distribution percentages categorized by molecular weight. (c) and (d) display the distribution percentages classified by molecular structure.
-
Specific name *GAU No. **NCBI No. Germination percentage after 12 h Spore
shapeSpore size (µm) ***Final germination length (µm) End of germination
time (h)Germination speed (μm/h) Length width Trichoderma atroviride 6022 MG807425.1 98.6 ± 0.40 ab Ellipsoidal 3.979 ± 0.059 hijk 4.125 ± 0.057 d 5.968575 8.761 ± 0.092 e 0.681 T. harzianum Ah90 KC576649.1 99.2 ± 0.37 ab Oval 3.177 ± 0.057 ijk 3.152 ± 0.060 e 4.764825 8.884 ± 0.071 e 0.536 T. virens 6011 KP671477.1 99.4 ± 0.40 a Oval 2.626 ± 0.029 k 2.595 ± 0.029 h 3.938745 11.800 ± 0.109 c 0.334 Penicillium implicatum MK-RSB19 OP411018.1 99.0 ± 0.01 ab Oval 2.852 ± 0.043 jk 2.905 ± 0.039 f 4.277535 14.780 ± 0.108 b 0.289 P. italicum MK-RSB20 OP411019.1 99.0 ± 0.45 ab Spherical 4.059 ± 0.055 hijk 4.149 ± 0.059 d 6.088485 14.823 ± 0.100 b 0.411 P. expansum MK-RSB18 OP411020.1 98.6 ± 0.40 ab Spherical 4.088 ± 0.062 hijk 4.084 ± 0.056 d 6.13221 14.823 ± 0.100 b 0.414 P. glabrum MK-RSB26 OP411021.1 99.2 ± 0.37 ab Spherical 2.884 ± 0.044 jk 2.912 ± 0.043 f 4.325355 11.782 ± 0.077 c 0.367 P. digitatum MK-RSB10 OP411022.1 98.8 ± 0.20 ab Spherical 2.824 ± 0.045 jk 2.834 ± 0.042 gf 4.236135 14.784 ± 0.051 b 0.287 Aspergillus niger MK-RSB7 OP411015.1 99.0 ± 0.32 ab Oval 4.317 ± 0.069 hij 4.050 ± 0.060 d 6.476145 10.190 ± 0.054 d 0.636 A. flavus MK-RSB28 OP411016.1 99.0 ± 0.45 ab Spherical 4.364 ± 0.072 hij 4.142 ± 0.059 d 6.54576 10.019 ± 0.207 d 0.653 Botrytis cinerea MK-RSB24 OP411017.1 98.8 ± 0.37 ab Ellipsoidal 7.934 ± 0.106 g 8.161 ± 0.116 b 11.90137 14.804 ± 0.066 b 0.804 Cladosporium cladosporioides pc4 MK765911.1 99.0 ± 0.01 ab Cylindrical 15.536 ± 0.278 e 2.587 ± 0.028 h 23.30463 14.718 ± 0.039 b 1.583 C. ramotenellum AM55 MH259170.1 99.4 ± 0.40 a Oval 12.399 ± 0.132 f 4.112 ± 0.052 d 18.59868 22.099 ± 0.219 a 0.842 C. limoniforme Br15 MH245072.1 98.4 ± 0.24 ab Cylindrical 4.655 ± 0.083 hi 1.802 ± 0.042 j 6.9828 14.790 ± 0.100 b 0.472 C. tenuissimum K15 MH258971.1 99.2 ± 0.49 ab Cylindrical 5.137 ± 0.055 h 2.092 ± 0.055 i 7.70484 14.759 ± 0.077 b 0.522 Fusarium oxysporum 7391 MK790682.1 98.8 ± 0.37 ab Fusiform 8.765 ± 0.198 g 3.141 ± 0.055 e 13.14744 14.717 ± 0.091 b 0.893 F. proliferatum pc91 MK765917.1 99.0 ± 0.32 ab Fusiform 19.703 ± 0.317 d 2.667 ± 0.028 hg 29.55448 14.806 ± 0.066 b 1.996 F. solani pc13 MK765916.1 98.2 ± 0.21 b Fusiform 13.612 ± 0.347 f 4.112 ± 0.059 d 20.41795 14.895 ± 0.049 b 1.371 Alternaria alternata MK-RSB6 OP411012.1 99.2 ± 0.20 ab Obclavate 49.795 ± 1.761 a 8.582 ± 0.199 a 74.69284 11.680 ± 0.049 c 6.395 A. tenuissima MK-RSB4 OP411013.1 99.2 ± 0.23 ab Obclavate 29.929 ± 0.594 c 7.640 ± 0.145 e 44.89369 14.754 ± 0.090 b 3.043 A. arborescens MK-RSB9 OP411014.1 98.6 ± 0.24 ab Obclavate 34.556 ± 0.907 b 7.738 ± 0.145 e 51.83403 14.923 ± 0.077 b 3.473 Replicate 5 100 100 5 Sum of squares 10.133 317496.495 8442.868 855.426 df 20 20 20 20 Mean square 0.507 15874.825 422.143 42.771 F-value 0.917 711.693 645.758 845.531 p-value 0.567 0.0001 0.0001 0.0001 * Accession number of culture collection of agricultural microorganisms at Gorgan University of Agricultural Science and Natural Resources. ** Accession number of National Center for Biotechnology Information. *** One and a half times the spore length was considered as the final germination length and growth of the tube before branching and turning into the mycelia. The lowercase letters indicate groups based on Duncan's multiple range test. Table 1.
Spore characteristics of fungal isolates.
-
Pathogenic post-harvest fungus Analysis section Sum of squares df Mean square F-value p-value Penicillium implicatum (A) 25.479 2 12.740 3.746 0.054 P. italicum (B) 66.173 2 33.087 8.890 0.004 P. expansum (C) 592.420 2 296.210 89.211 0.0001 P. glabrum (D) 61.142 2 30.571 9.339 0.004 P. digitatum (E) 114.723 2 57.361 18.508 0.0001 Aspergillus niger (F) 1,178.349 2 589.174 191.312 0.0001 A. flavus (G) 10.646 2 5.323 1.557 0.250 Botrytis cinerea (H) 666.296 2 333.148 113.961 0.0001 Cladosporium cladosporioides (I) 166.026 2 83.013 26.223 0.0001 C. ramotenellum (J) 800.174 2 400.087 120.219 0.0001 C. limoniforme (K) 993.039 2 496.519 159.835 0.0001 C. tenuissimum (L) 878.684 2 439.342 147.069 0.0001 Fusarium oxysporum (M) 49.905 2 24.952 6.971 0.010 F. proliferatum (N) 341.754 2 170.877 49.327 0.0001 F. solani (O) 98.872 2 49.436 16.515 0.0001 Alternaria alternata (P) 373.321 2 186.660 54.973 0.0001 A. tenuissima (Q) 12.768 2 6.384 2.014 0.176 A. arborescens (R) 43.295 2 21.648 6.818 0.011 Not significant: p-value > 0.05; Significant: p-value < 0.05. Table 2.
Results of one-way analysis of variance (ANOVA) for duncan's multiple range test mean comparisons in Fig. 2.
-
Peak no. Metabolite Time Area % Area Molecular formula Molecular weight (g/mol) % Match against NIST* CAS** registration no. 1 Pentanal 11.086 808690 0.8 C5H10O 86.134 78 110-62-3 2 2-Hexanone, 5-methyl 11.164 108673 0.11 C7H14O 114.185 78 110-12-3 3 Tridecane 14.64 13189397 12.97 C13H28 184.36 74 629-50-5 4 Dodecane 18.537 1456396 1.43 C12H26 170.34 94 112-40-3 5 Sulfurous acid, 2-propyl tridecyl ester 25.308 3418750 3.36 C19H40O3S 348.59 95 309-12-4 6 4-Methylcyclohexanol 27.42 2268233 2.23 C7H14O 114.19 82 589-91-3 7 4-Methylheptane-3,5-dione 27.918 1917646 1.89 C8H14O2 142.2 77 1187-04-8 8 5-Tridecanone 28.136 328445 0.32 C13H26O 198.34 76 30692-16-1 9 Lauric acid 28.281 492818 0.48 C12H24O2 200.32 83 143-07-7 10 Margaric acid 28.38 702940 0.69 C17H34O2 270.46 82 506-12-7 11 Pentane, 1-butoxy 28.427 254993 0.25 C9H20O 144.26 88 18636-66-3 12 Cytidine 28.551 429770 0.42 C9H13N3O5 243.22 76 65-46-3 13 1,5-anhydro-arabino-furanose 28.748 1292085 1.27 C5H6O3 114.1 77 51246-91-4 14 Phenol, 2,4-bis(1,1-dimethylethyl) 28.878 3820885 3.76 C14H22O 206.33 96 96-76-4 15 Hexadecane 31.306 2198992 2.16 C16H34 226.44 96 544-76-3 16 1,1-Bis(p-tolyl)ethane 34.347 1348729 1.33 C20H22 262.39 91 98211-18-9 17 Octadecane 36.703 917085 0.9 C18H38 254.51 97 593-45-3 18 Palmitic acid, methyl ester 39.873 3225855 3.17 C17H34O2 270.46 99 112-39-0 19 Benzenepropanoic acid, 3,5-bis(1,1-dimethylethyl)-4-hydroxy-, methyl ester 40.278 6285759 6.18 C23H36O3 360.53 99 6386-38-5 20 Palmitic acid 40.822 6825347 6.71 C16H32O2 256.42 99 57-10-3 21 Eicosane 41.603 859015 0.84 C20H42 282.56 94 112-95-8 22 Tetracosane 46.085 946504 0.93 C24H50 338.65 90 646-31-1 23 1,4-Benzenedicarboxylic acid, bis(2-ethylhexyl) ester 53.106 48578756 47.78 C24H38O4 390.62 91 6422-86-2 * National Institute of Standards and Technology (version 23). ** Chemical Abstracts Service. Table 3.
Details of the identified metabolites from initial spore germination stage of Trichoderma atroviride.
-
Peak no. Metabolite Time Area % Area Molecular formula Molecular weight (g/mol) % Match against NIST* CAS**
registration no.1 2-Hexanone, 5-methyl 11.045 1308158 0.73 C7H14O 114.19 80 110-12-3 2 Propene, 1,1'-oxybis 11.081 140978 0.08 C6H12O 100.16 78 4696-29-1 3 5-Methyluracil 14.666 14678214 8.19 C5H6N2O2 126.11 80 65-71-4 4 Dodecane 18.532 1324048 0.74 C12H26 170.34 94 112-40-3 5 Tetradecane 25.303 3020121 1.68 C14H30 198.41 94 629-59-4 6 d-Lyxo-d-manno-nononic-1,4-lactone 27.322 559198 0.31 C9H16O9 268.22 72 3080-49-7 7 5-Tridecanone 27.467 515994 0.29 C13H26O 198.34 74 30692-16-1 8 1,5-anhydro-arabino-furanose 27.591 402534 0.22 C5H6O3 114.1 83 51246-91-4 9 Undecanoic acid 28.38 706513 0.39 C11H22O2 186.29 87 112-37-8 10 Pentane, 1-butoxy 28.458 106359 0.06 C9H20O 144.26 83 18636-66-3 11 2,4-Di-tert-butylphenol 28.51 37983 0.02 C14H22O 206.32 83 96-76-4 12 Decanoic acid 28.878 4084001 2.28 C10H20O2 172.26 96 112-37-8 13 Hexadecane 31.306 2051964 1.14 C16H34 226.44 96 544-76-3 14 1,1-Bis(p-tolyl)ethane 34.347 1583496 0.88 C16H18 210.31 90 98211-18-9 15 Octadecane 36.703 762932 0.43 C18H38 254.51 95 593-45-3 16 Hexadecanoic acid, methyl ester 39.868 2090101 1.17 C17H34O2 270.45 98 112-39-0 17 Benzenepropanoic acid, 3,5-bis(1,1-dimethylethyl)-4-hydroxy-, methyl ester 40.278 6308220 3.52 C14H20O3 236.31 99 6386-38-5 18 Palmitic acid 40.796 2276043 1.27 C16H32O2 256.42 99 57-10-3 19 2-methyloctacosane 46.084 838744 0.47 C29H60 408.8 90 1560-98-1 20 1,4-Benzenedicarboxylic acid, bis(2-ethylhexyl) ester 53.138 128988610 71.94 C24H38O4 390.62 91 422-86-2 21 Phthalic acid, di(2-propylphenyl) ester 56.669 7520123 4.19 C26H26O4 402.5 94 98661-89-0 * National Institute of Standards and Technology (version 23). ** Chemical Abstracts Service. Table 4.
Details of the identified metabolites from initial spore germination stage of Trichoderma virens.
-
Peak no. Metabolite Time Area % Area Molecular formula Molecular weight (g/mol) % Match against NIST* CAS** registration no. 1 2,4-Diamino-6-hydroxypyrimidine 14.614 3064400 3.39 C4H6N4O 126.12 64 56-06-4 2 Dodecane 18.532 1314498 1.45 C12H26 170.34 95 112-40-3 3 Tetradecane 25.303 2574175 2.84 C14H30 198.41 94 629-59-4 4 2-Vinyl-9-[beta-d-ribofuranosyl]hypoxanthine 27.446 2051010 2.27 C12H14N4O5 294.26 77 110851-56-4 5 5-Tridecanone 27.524 209944 0.23 C13H26O 198.34 75 30692-16-1 6 1,5-anhydro-arabino-furanose 27.737 473868 0.52 C5H6O3 114.1 72 51246-91-4 7 Itaconic acid 27.788 85858 0.09 C5H6O4 130.09 85 97-65-4 8 Cyclopentanol 27.892 96554 0.11 C5H10O 86.1323 87 96-41-3 9 2,4-Di-tert-butylphenol 28.878 1984097 2.19 C14H22O 206.32 96 96-76-4 10 Hexadecane 31.306 1866061 2.06 C16H34 226.44 98 544-76-3 11 1,1-Bis(p-tolyl)ethane 34.347 1533054 1.69 C16H18 210.31 91 98211-18-9 12 Octadecane 36.697 1005421 1.11 C18H38 254.51 96 593-45-3 13 Hexadecanoic acid, methyl ester 39.873 2703467 2.99 C17H34O2 270.45 98 112-39-0 14 Benzenepropanoic acid, 3,5-bis(1,1-dimethylethyl)-4-hydroxy-, methyl ester 40.277 6624034 7.32 C14H20O3 236.31 99 6386-38-5 15 Palmitic acid 40.802 2434738 2.69 C16H32O2 256.42 99 57-10-3 16 Icosane 41.606 2069726 2.29 C20H42 282.54 97 112-95-8 17 2-methyloctacosane 46.089 1568284 1.73 C29H60 408.8 98 1560-98-1 18 Heneicosane, 11-(1-ethylpropyl) 50.214 846188 0.94 C26H54 366.7 87 55282-11-6 19 Phthalic acid, di(2-propylpentyl) ester 53.088 13204422 14.59 C26H26O4 402.5 91 998661-89-0 20 1,4-Benzenedicarboxylic acid, bis(2-ethylhexyl) ester 56.684 44774953 49.48 C24H38O4 390.62 94 6422-86-2 * National Institute of Standards and Technology (version 23). ** Chemical Abstracts Service. Table 5.
Details of the identified metabolites from initial spore germination stage of Trichoderma harzianum.
Figures
(3)
Tables
(5)