-
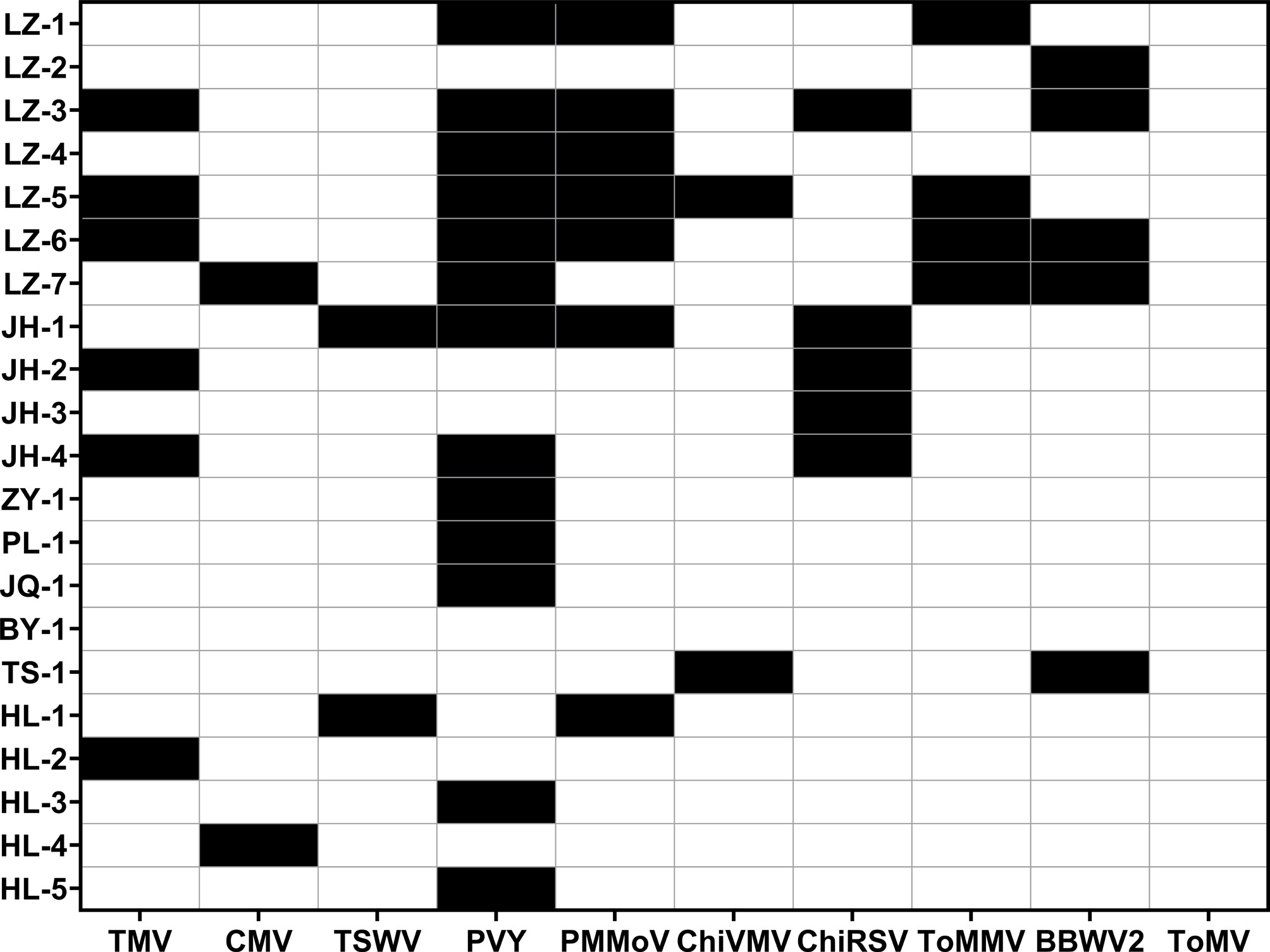
Figure 1.
Viral diseases detection results of 21 samples. Black boxes signify the detection of viral diseases, whereas the white boxes indicate none.
-
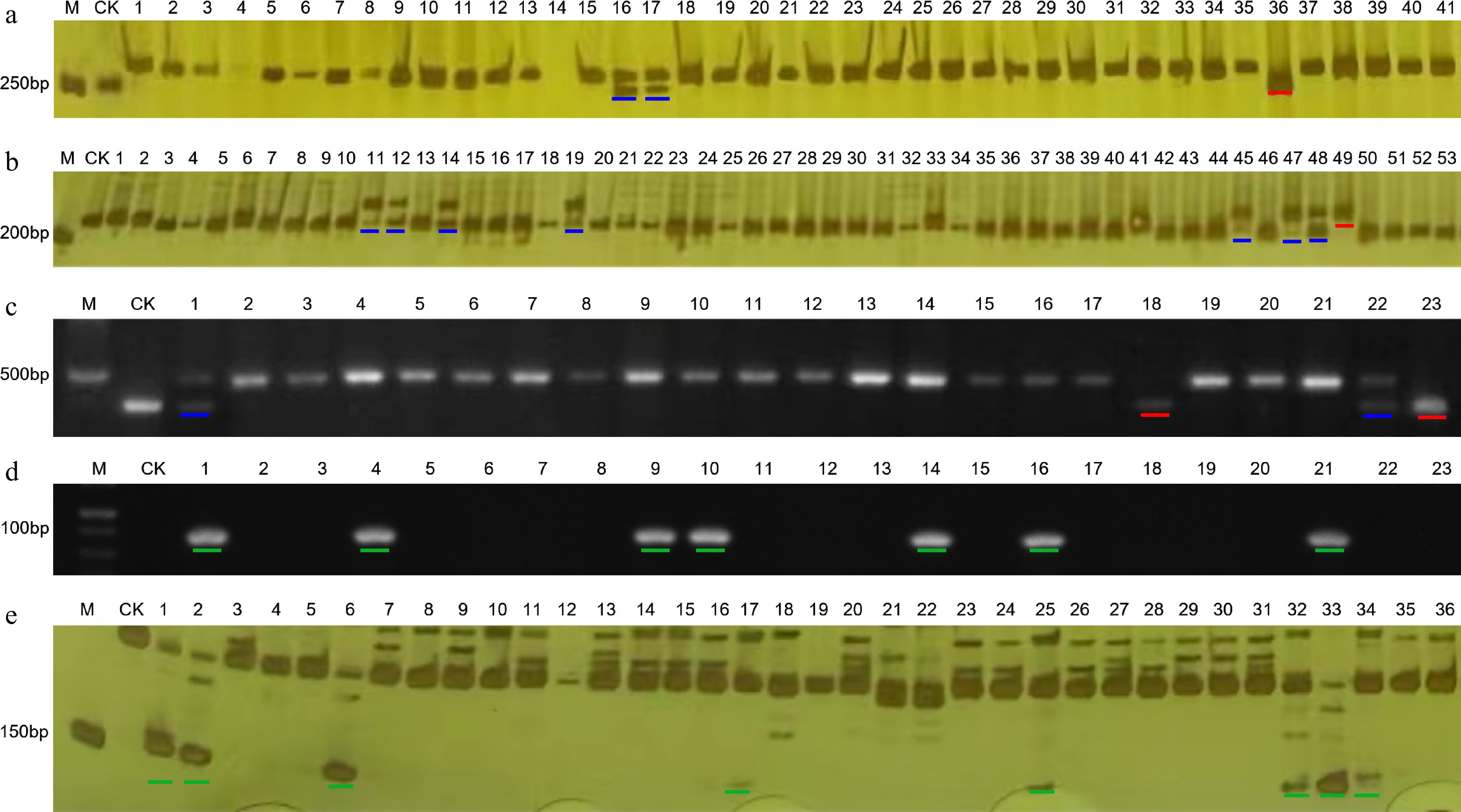
Figure 2.
The genotyping results of some germplasms obtained by molecular markers. (a) Marker PMFR11283. (b) InDel-2-134 marker. (c) Marker PVY-CAPS. (d) Marker L4-SCAR. (e) Marker Chr-lcv-7. M: DNA marker; CK: negative control. The red line indicates a homozygous genotype for resistance to disease, while the blue line indicates a heterozygous genotype for resistance to disease. The green lines represent the disease-resistant genotypes indicated by dominant markers.
-

Figure 3.
Artificial inoculation of germplasm accessions resistant to TMV and CMV in peppers. (a) Phenotypes of plants inoculated with TMV for 25 d. (b) Phenotypes of plants inoculated with CMV for 25 d.
-
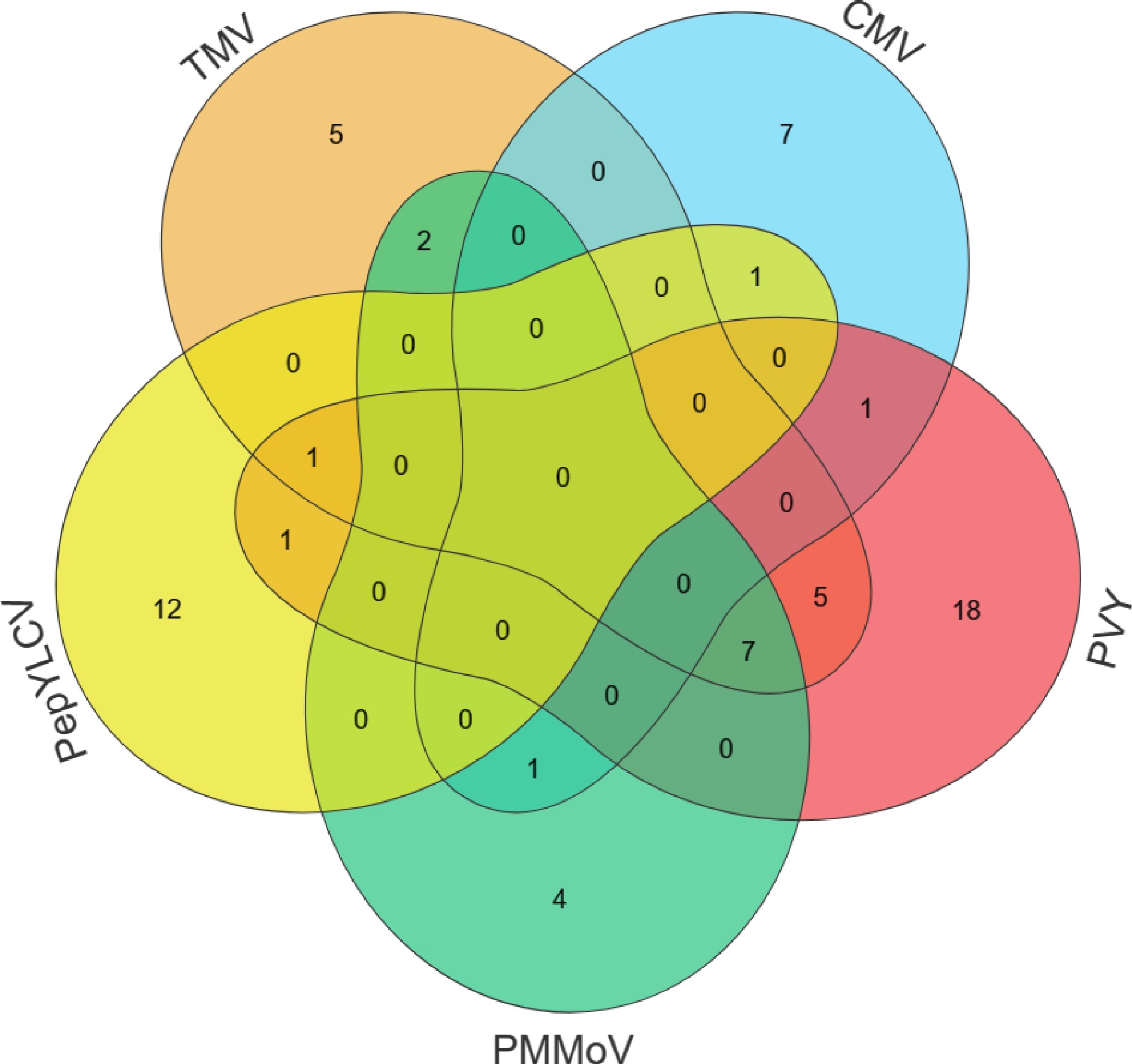
Figure 4.
Statistics of the number of five virus-resistant germplasms.
-
Virus Forward sequence (5'–3') Reverse sequence (5'–3') Fragment size
(bp)Annealing
temperature (°C)Ref. CMV TCTCATGGATGCTTCTCCGCG CCGTAAGCTGGATGGACAACC 760 55 [32] ChiVMV AAGATATGGGCTTCAAGAAACCTTACCG CCTACCCTACCGTCCAGTCCGAACATCCT 174 58 [33] ChiRSV ATGAACTGTACGCCGAAGGG AGTCCGATCGCCTATGAGGA 897 53 [34] TMV GTTCTTGTCATCTGCGTGGG GCTCTCGAAAGAGCTCCGAT 406 55 [34] TSWV TCACTGTAATGTTCCATAGCAA AGAGCAATYGTGTCAATTTTATTC 861 52 [32] ToMV GTTTCATTGTGCTGTTGAGTAC AGAAGTACCCATATTGCTTCTTG 362 57 [35] ToMMV ATGTCTTDCGCTATTACTTCTCCG CTCTGGTTGTAGAAACCTGTT 480 56 [35] PVY GGCATACGGACATAGGAGAAACT CTCTTTGTGTTCTCCTCTTGTGT 447 57 [36] PMMoV ATGGCTTACACAGTTTCC TTAAGGAGTTGTAGCCCAG 474 57 [37] BBWV2 AGTTATATGCTTGGGCAAGCGCATG CATGAACATTCCCCATCTCCACGTG 490 57 [32] Table 1.
Primers for identification of 10 viruses.
-
Virus Marker ID Forward primer (5'–3') Reverse primer (5'–3') Ref. TMV PMFR11283 CTGCAGAACAACAATGGCACG GGACTGCAGAGGAGGAAGC [23] CMV InDel-2-134 TGCTTCAGTTGAGTTGTCCA TAAATCCCCTTGTGGTGGCT [29] PVY PVY-CAPS CGAAGAGAGAAGGTC TCAGGGTAGGTTATT [24] PMMoV L4-SCAR ATCGATGCACCCCTCGTTTTAATC GAGCAGTGTGGAGTGTCTATTGCTCA [30] PepYLCV Chr-lcv-7 CTGATAACTGACAGTTTAGATAGGAATTGG CAACTCAGTCTATAACCGGTGTATG [31] Table 2.
Marker information for identification of resistance to virus disease in pepper germplasm.
-
Resistance Disease index (DI) Immune (I) DI is 0 Highly resistant (HR) DI is greater than 0 and less than 10 Resistant (R) DI is greater than or equal to 10 and less than 20 Moderately resistant (MR) DI is greater than or equal to 20 and less than 40 Susceptible (S) DI is greater than or equal to 40 and less than 60 Highly susceptible (HS) DI is greater than or equal to 60 and less than or equal to 100 Table 3.
Evaluation criteria of resistance to TMV and CMV in pepper.
-
TMV (PMFR11283) CMV (InDel-2-134) PVY (PVY-CAPS) PMMoV (L4-SCAR) PepYLCV (Chr-lcv-7) A98, SP14, SP16, SP18, SP21, SP27, SP39, SP41, SP44,
SP45, SP46, SP48, SP55,
SP56, SP57, SP64, SP108, SP113, Suxuan 1,
Shenglong 4A33, SP34, SP35, SP37, SP42, SP68, SP70, SP71, SP72, SP79 A43, A45, A83, A88, SP14, SP16, SP19, SP21, SP36, SP37, SP38, SP41, SP44, SP45, SP46, SP47, SP48, SP50, SP51, SP53, SP54, SP55, SP56, SP62, SP64, SP65, SP66, SP93, SP94, SP96, SP97, SP108, SP113 A70, A78, SP14, SP16, SP21, SP40, SP48, SP55, SP56,
SP57, SP64, SP71, M4,
Suxuan 1A4, A22, A80, A89, A92, A97, SP35, SP51, SP90, SP106, SP108, R3, R8, R10, R11 Table 4.
Statistics of germplasm accessions with resistance to viral diseases in pepper.
-
Category Pattern Germplasm accessions Single TMV A98, SP18, SP27, SP39, Shenglong 4 CMV A33, SP34, SP42, SP68, SP70, SP72, SP79 PVY A43, A45, A83, A88, SP19, SP36, SP38, SP47, SP50, SP53, SP54, SP62, SP65, SP66, SP93, SP94, SP96, SP97 PMMoV A70, A78, M4, SP40 PepYLCV A4, A22, A80, A89, A92, A97, R10, R11, R3, R8, SP106, SP90 Double TMV + PMMoV SP57, Suxuan 1 TMV + PVY SP41, SP44, SP45, SP46, SP113 PVY + PepYLCV SP51 CMV + PepYLCV SP35 CMV + PMMoV SP71 CMV + PVY SP37 Triple TMV + PVY + PepYLCV SP108 TMV + PVY + PMMoV SP14, SP16, SP21, SP48, SP55, SP56, SP64 Table 5.
Statistics on pepper germplasm accessions resistant to viral diseases.
Figures
(4)
Tables
(5)