-
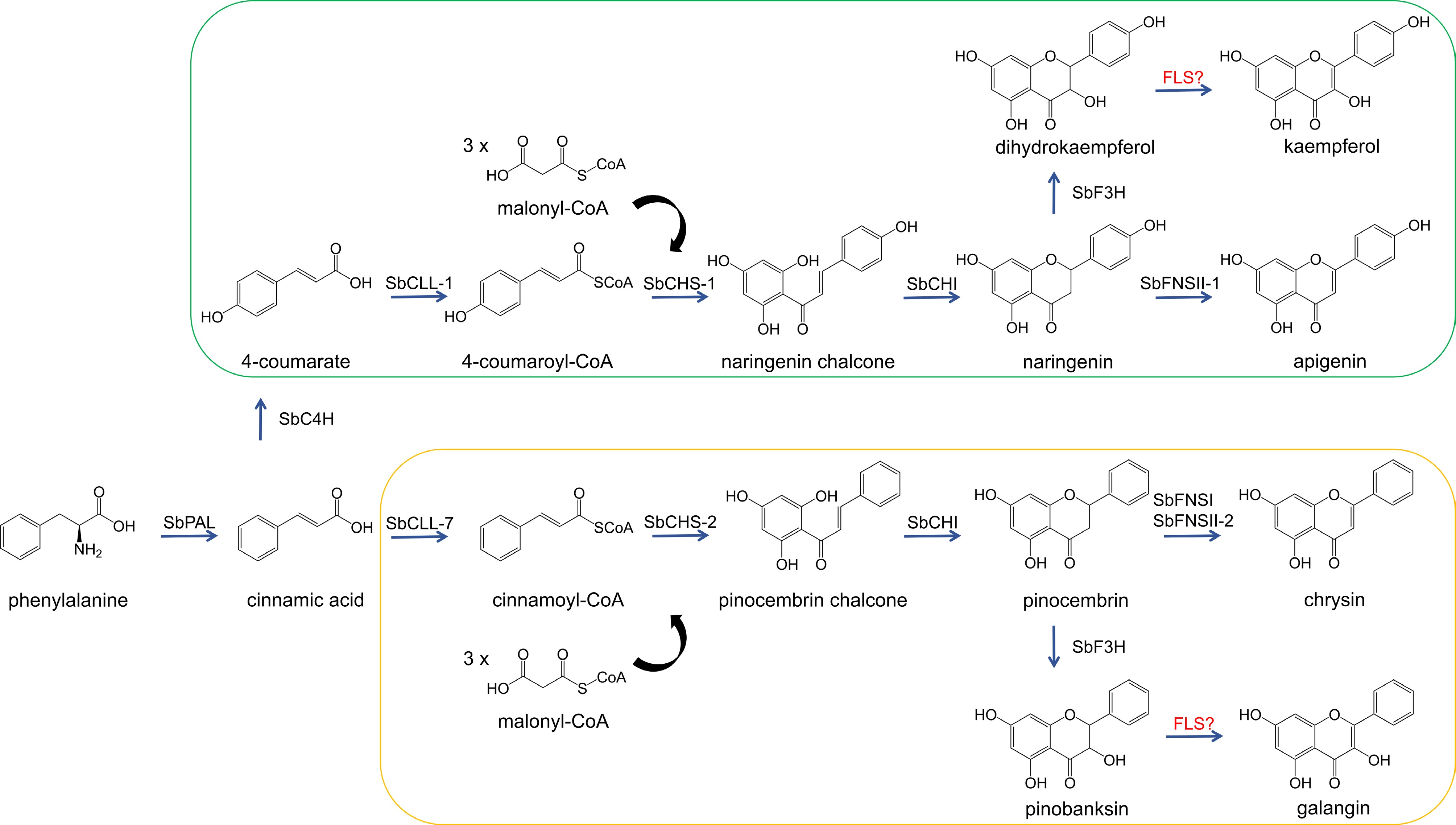
Figure 1.
The biosynthetic pathway of flavonols in S. baicalensis. SbPAL, phenylalanine ammonialyase; SbC4H, cinnamate 4-hydroxylase; SbCLL-1, 4-coumarate CoA ligase; SbCHS, chalcone synthase; SbCHI, chalcone isomerase; SbFNS, flavone synthase; SbCLL-7, cinnamate-CoA ligase; SbF3H, flavanone 3-hydroxylase; FLS, flavonol synthase. The enzymes highlighted in red are still under investigation. The green-bordered part shows the metabolic pathway of 4'-hydroxyflavonoids; the yellow-bordered part shows the metabolic pathway of 4'-deoxyflavonoids.
-
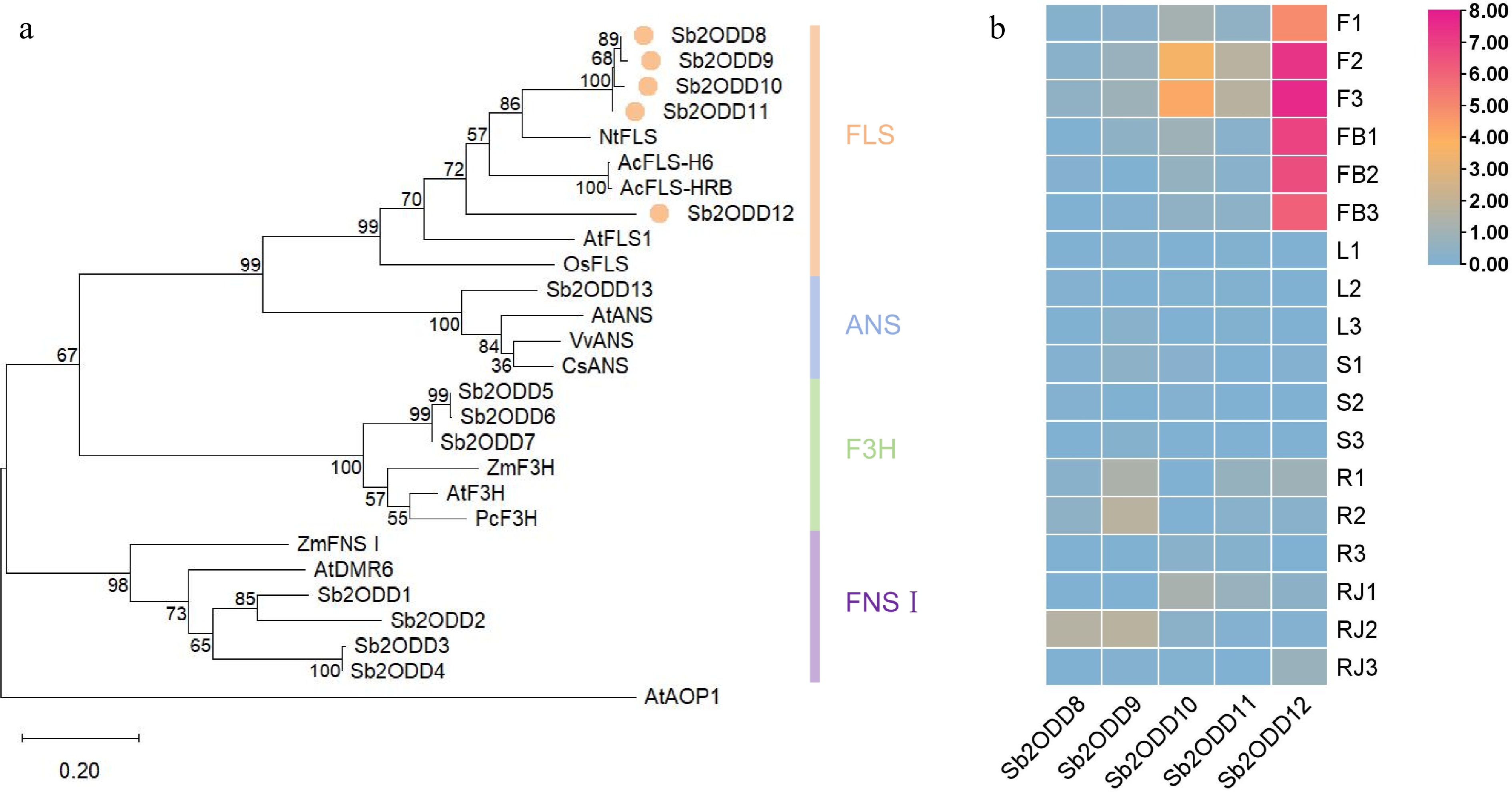
Figure 2.
Phylogenetic analysis and expression patterns of Sb2ODDs in S. baicalensis. (a) Phylogenetic tree of the 2ODD family members involved in flavonoid biosynthesis. The maximum-likelihood method was used to construct this tree with 1,000 replicate bootstrap support. The tree was rooted with AtAOP1. Accession numbers of the proteins used and their species names: NtFLS1, ABE28017, Nicotiana tabacum; AcFLS-H6, KY369209, Allium cepa; AcFLS-HRB, KY369210, Allium cepa; AtFLS1, At5g08640, Arabidopsis thaliana; OsFLS1, NP_001048230, Oryza sativa; AtANS, At4g22880; VvANS, ABM67590, Vitis vinifera; CsANS, AAT02642, Citrus sinensis; ZmF3H, NP_001130275, Zea mays; AtF3H, NP_190692; PcF3H, AAP57394, Petroselinum crispum; ZmFNSI, NP_001151167; AtDMR6, NP_197841; Sb2ODD1, Sb06g22120, Scutellaria baicalensis; Sb2ODD2, Sb02g38930; Sb2ODD3, Sb01g51050; Sb2ODD4, Sb01g51220; Sb2ODD5, Sb05g01401; Sb2ODD6, Sb05g01404; Sb2ODD7, Sb05g01631; Sb2ODD8, Sb0g04190; Sb2ODD9, Sb05g15130; Sb2ODD10, Sb05g15090; Sb2ODD11, Sb05g11030; Sb2ODD12, Sb05g11050; Sb2ODD13, Sb03g04100. (b) Tissue-specific expression heatmap of Sb2ODD8–Sb2ODD12. The color scale on the right represents the FPKM values normalized with log10. F, flower; FB, flower bud; L, leaf; S, stem; R, root; RJ, MeJA-treated root. the numbers behind indicated biological replicates.
-
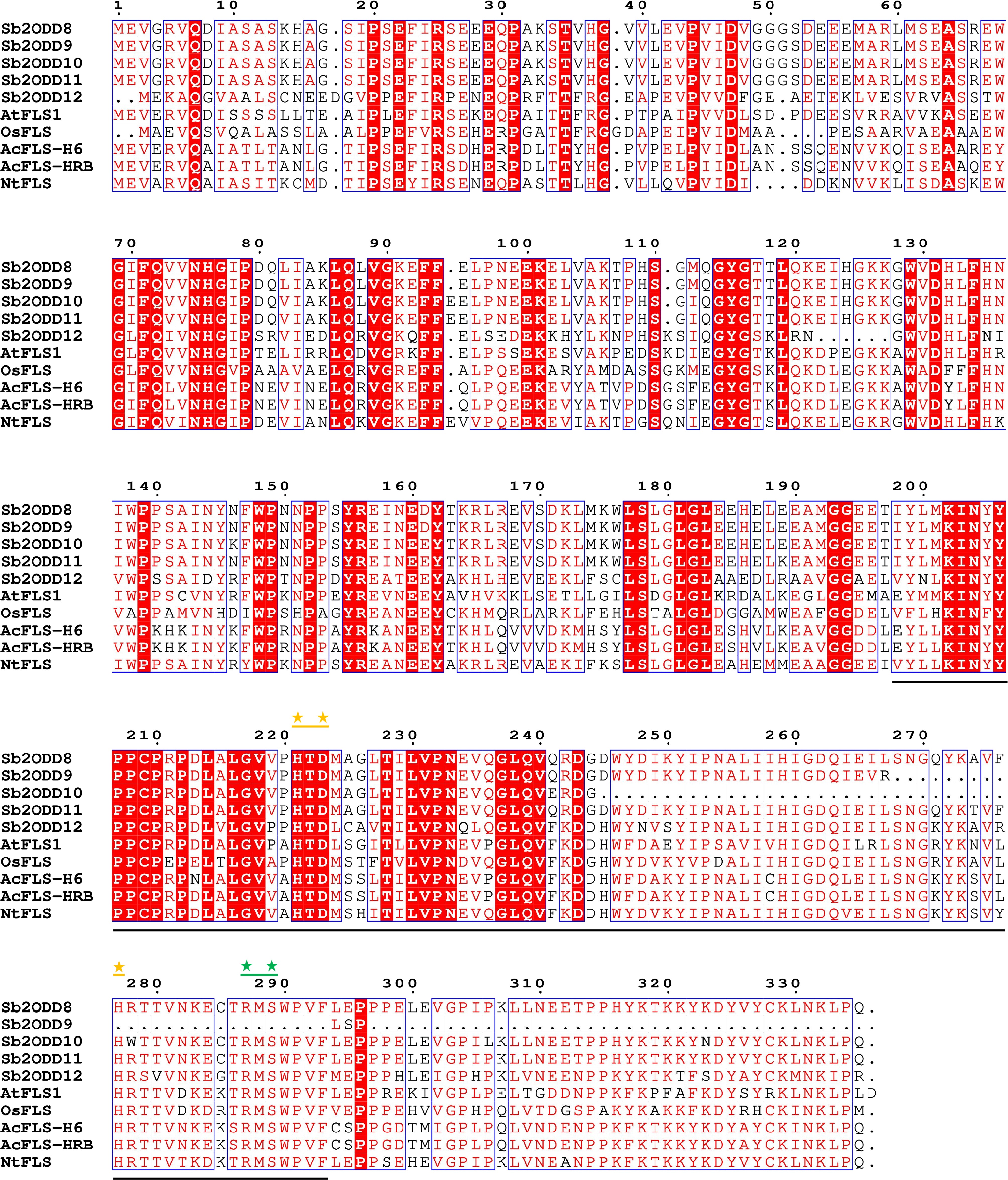
Figure 3.
Sb2ODDs and other species FLS sequences alignment. Underlined regions indicates conserved domains which coordinate Fe(II) binding and 2-oxoglutarate binding. Yellow asterisks mark amino acid residues important for Fe(II) binding. Green asterisks mark amino acid residues important for 2-oxoglutarate binding.
-
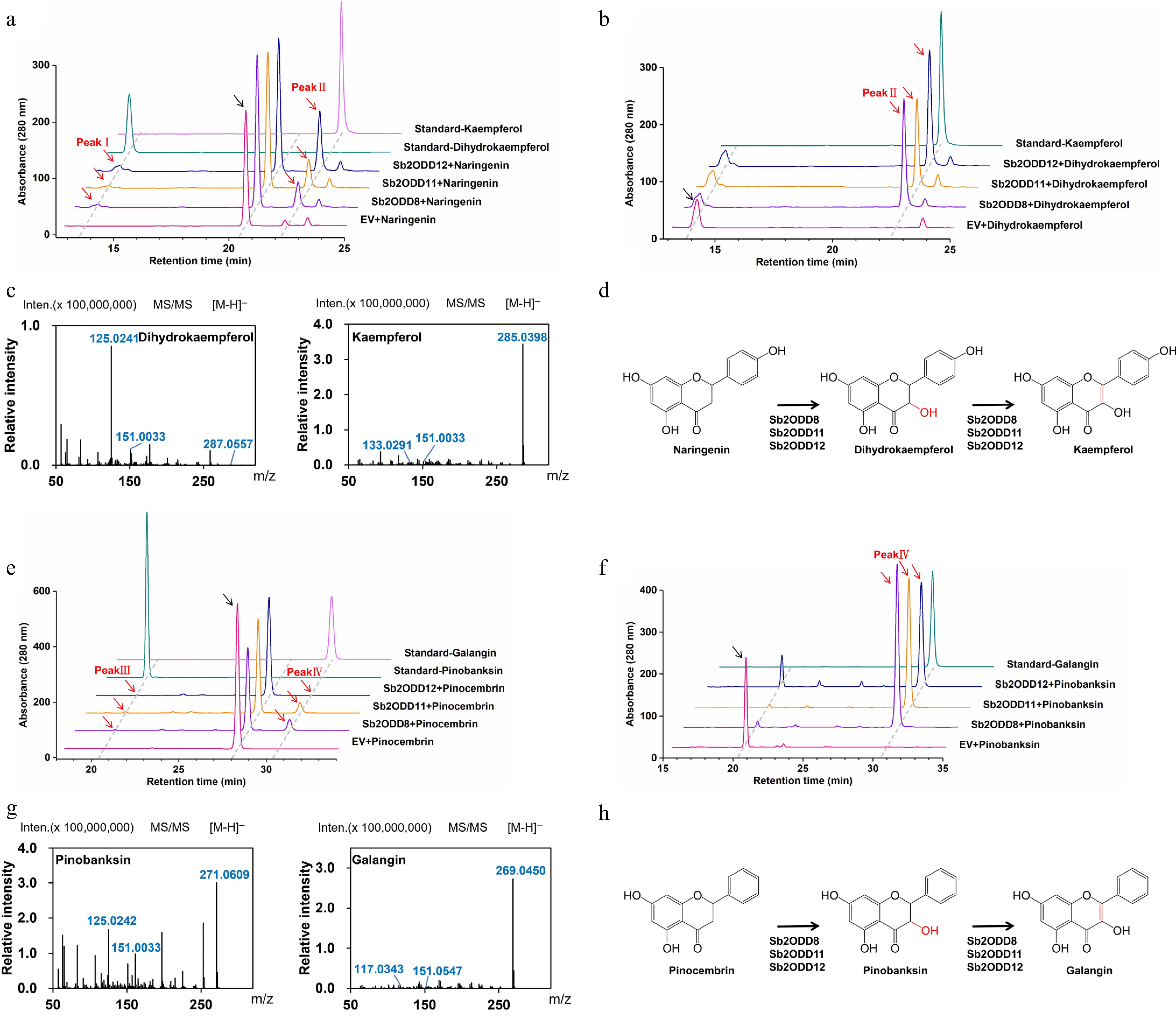
Figure 4.
In vivo yeast enzyme assays of Sb2ODDs. (a) Assays of activity in yeast in vivo. HPLC analysis of yeast samples incubated with naringenin: top, kaempferol standard; second, dihydrokaempferol standard; middle, Sb2ODD12, Sb2ODD11, Sb2ODD8 with naringenin; bottom, empty vector control (EV). (b) Assays of activity in yeast in vivo. HPLC analysis of yeast samples incubated with dihydrokaempferol: top, kaempferol standard; middle, Sb2ODD12, Sb2ODD11, Sb2ODD8 with dihydrokaempferol; bottom, empty vector control (EV). (c) MS/MS patterns of Sb2ODD12, Sb2ODD11, Sb2ODD8 products, which were identical to dihydrokaempferol and kaempferol standard. (d) The reaction was catalyzed by Sb2ODD12, Sb2ODD11, Sb2ODD8 using naringenin as a substrate. (e) Assays of activity in yeast in vivo. HPLC analysis of yeast samples incubated with pinocembrin: top, galangin standard; second, pinobanksin standard; middle, Sb2ODD12, Sb2ODD11, Sb2ODD8 with pinocembrin; bottom, empty vector control (EV). (f) Assays of activity in yeast in vivo. HPLC analysis of yeast samples incubated with pinobanksin: top, galangin standard; middle, Sb2ODD12, Sb2ODD11, Sb2ODD8 with pinobanksin; bottom, empty vector control (EV). (g) MS/MS patterns of Sb2ODD12, Sb2ODD11, Sb2ODD8 products, which were identical to pinobanksin and galangin standard. (h) The reaction was catalyzed by Sb2ODD12, Sb2ODD11, Sb2ODD8 using pinocembrin as a substrate.
-
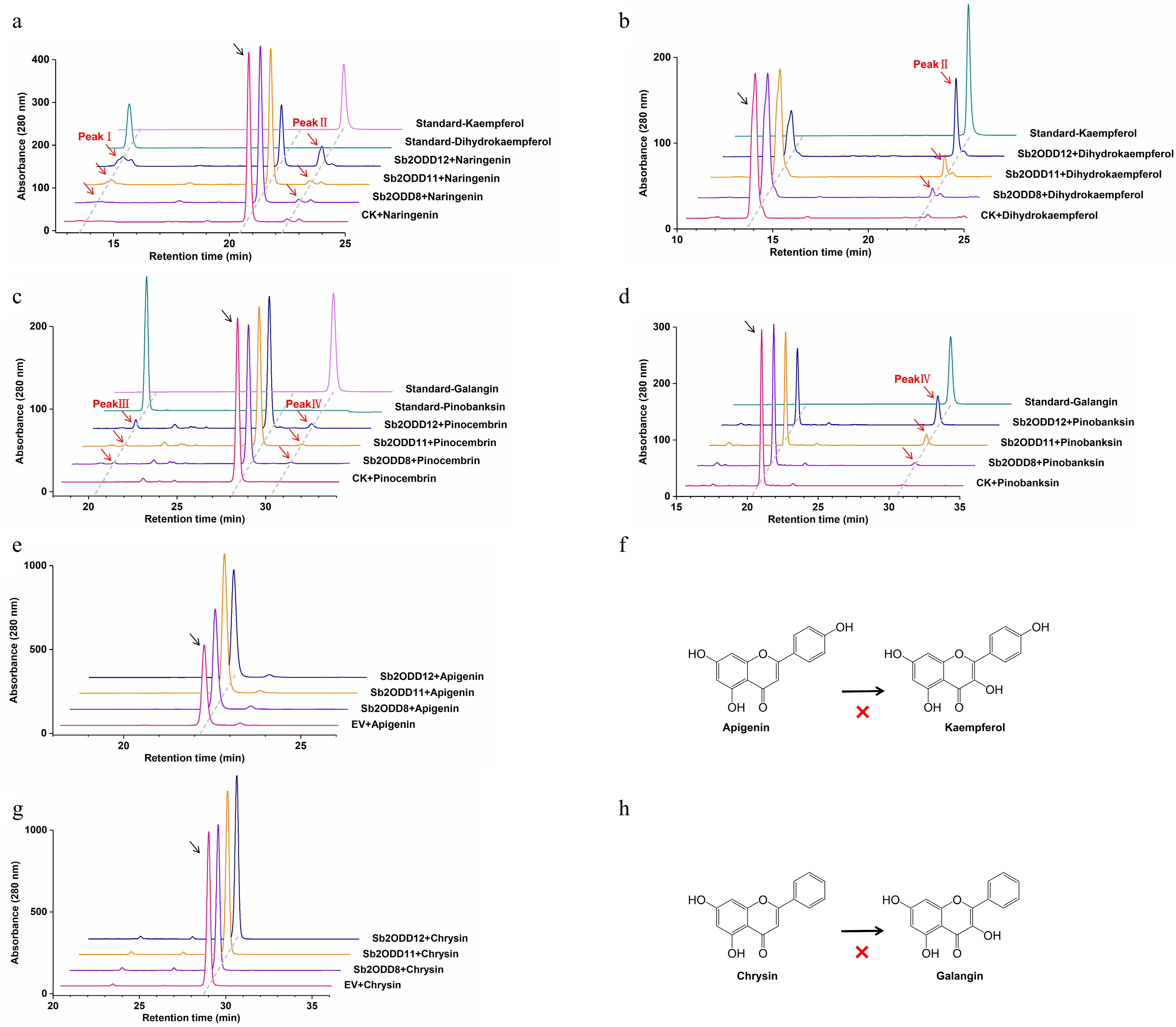
Figure 5.
In vitro enzyme assays of Sb2ODDs. (a) HPLC analysis of enzyme activity in vitro using naringenin as a substrate: top, kaempferol standard; second, dihydrokaempferol standard; middle, Sb2ODD12, Sb2ODD11, Sb2ODD8 with naringenin; bottom, control check (CK). (b) HPLC analysis of enzyme activity in vitro using dihydrokaempferol as a substrate: top, kaempferol standard; middle, Sb2ODD12, Sb2ODD11, Sb2ODD8 with dihydrokaempferol; bottom, control check (CK). (c) HPLC analysis of enzyme activity in vitro using pinocembrin as a substrate: top, galangin standard; second, pinobanksin standard; middle, Sb2ODD12, Sb2ODD11, Sb2ODD8 with pinocembrin; bottom, control check (CK). (d) HPLC analysis of enzyme activity in vitro using pinobanksin as a substrate: top, galangin standard; middle, Sb2ODD12, Sb2ODD11, Sb2ODD8 with pinobanksin; bottom, control check (CK). (e) HPLC analysis of samples incubated with apigenin: top to bottom are Sb2ODD12, Sb2ODD11, Sb2ODD8 and empty vector control (EV). (f) Sb2ODDs cannot catalyze the formation of kaempferol from apigenin. (g) HPLC analysis of samples incubated with chysin: top to bottom are Sb2ODD12, Sb2ODD11, Sb2ODD8 and empty vector control (EV). (h) Sb2ODDs cannot catalyze the formation of galangin from chysin.
-
Enzyme Substrate Vmax (pkat/mg) Km (μM) Kcat (S) KcatlKm (M/S) Sb2ODD8 Dihydrokaempferol 360.80 125.30 242.22 1.93 × 106 Pinobanksin 404.30 143.10 271.42 1.90 × 106 Sb2ODD11 Dihydrokaempferol 105.50 10.60 71.07 6.70 × 106 Pinobanksin 92.86 8.60 62.56 7.27 × 106 Sb2ODD12 Dihydrokaempferol 542.20 55.81 356.95 6.40 × 106 Pinobanksin 361.70 54.17 238.12 4.40 × 106 Table 1.
Kinetic analysis of Sb2ODDs.
Figures
(5)
Tables
(1)