-
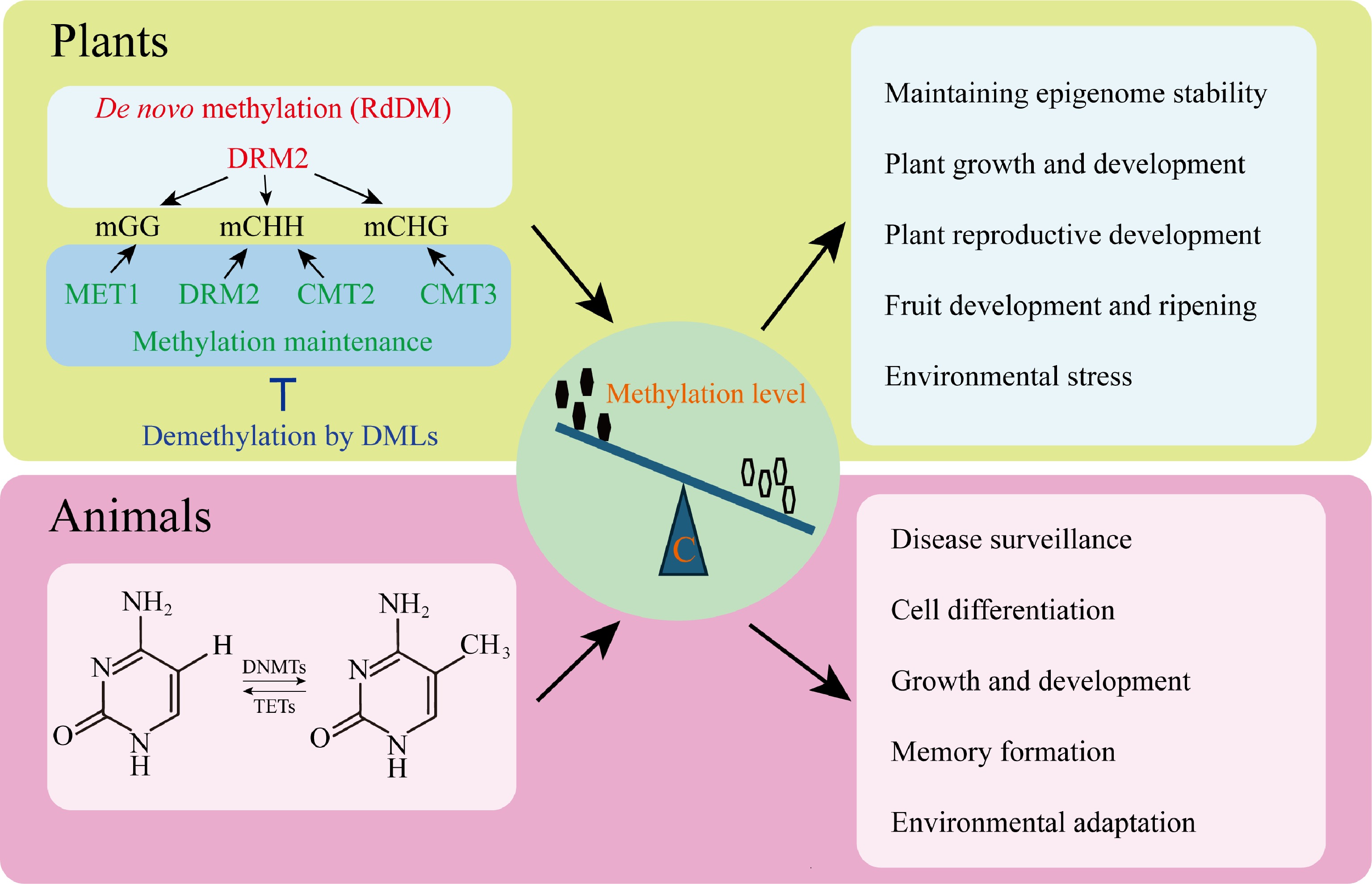
Figure 1.
Core regulatory mechanisms of DNA methylation in eukaryotes. Cytosine DNA methylation is self-regulated both during maintenance and de novo in plants. The RNA-directed DNA methylation (RdDM) pathway uses the DOMAINS REARRANGED METHYLTRANSFERASE TWO (DRM2) to recruit de novo methylation and METHYLTRANSFERASE1 (MET1) to maintain CG methylation. DNA methylation in plants is maintained by plant-specific CHROMOMETHYLASEs (CMTs), while CHH methylation is maintained by CMTs and DRM2, both independently and in conjunction with RdDM. It is counteracted by DNA demethylases such as REPRESSOR OF SILENCING ONE (ROS1), DEMETER (DME)-LIKE TWO (DML2), and DML3 that activate the expression of genes. The combination of these pathways ensures the stability of epigenetics and controls key biological functions, including plant development, reproduction, fruit development, and ripening, as well as the stress response. The CpG dinucleotides of DNA methylation are mainly confined to animals and regulated by the counteracting activities of DNA METHYLTRANSFERASEs (DNMTs) and ten-eleven translocation (TET) proteins. This control equilibrium supports the process of development, cell differentiation, defense against disease, formation of memory, and adaptation to the environment.
-
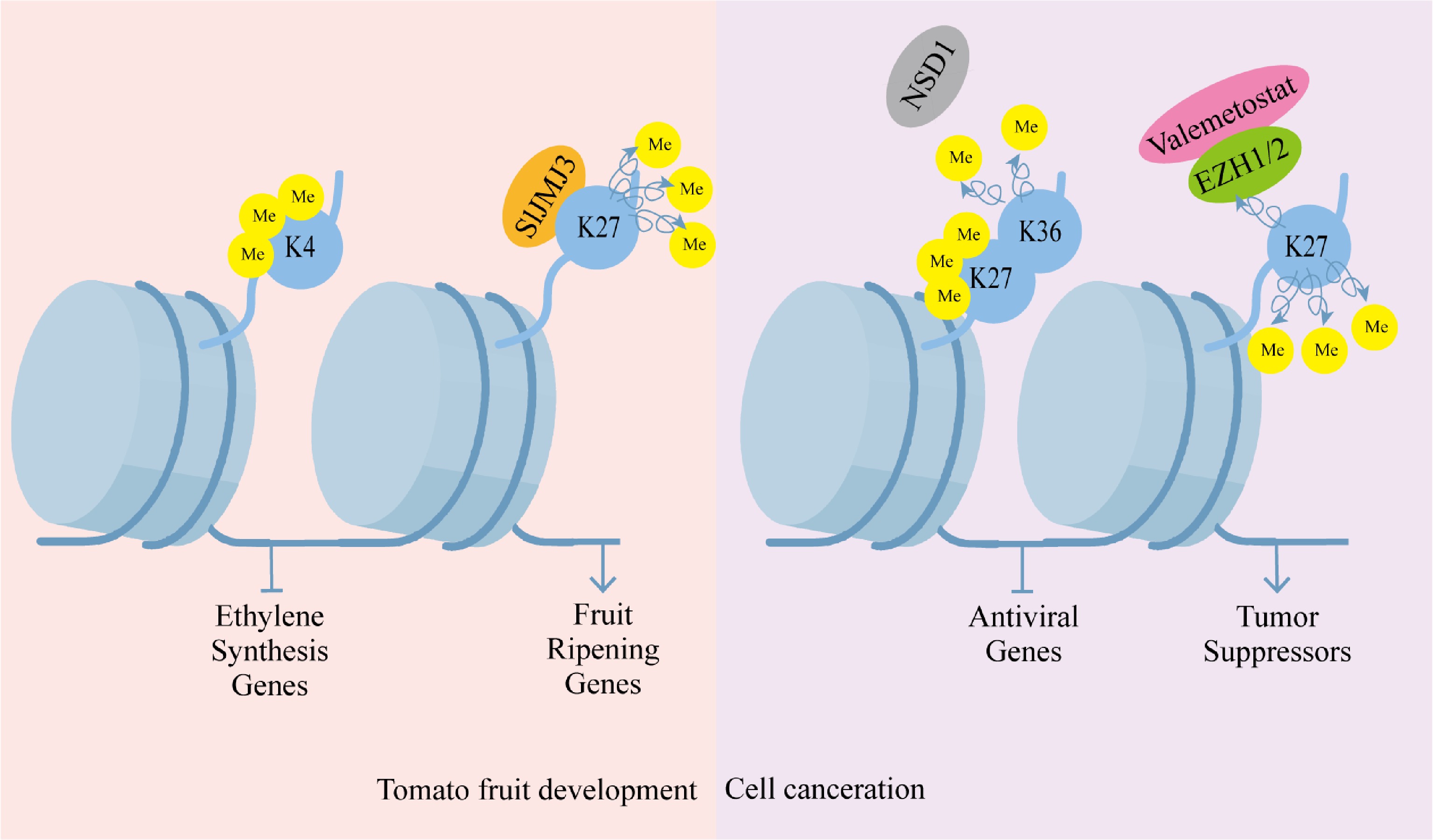
Figure 2.
Histone methylation regulation pattern. Histone methylation regulation is crucial in plant development. Histone H3 lysine four trimethylation (H3K4me3) is linked to the silencing of ethylene biosynthesis genes during tomato fruit ripening, and the histone demethylase Jumonji C domain-containing protein three (SlJMJ3) facilitates ripening by deposing the ethylene biosynthesis-related genes of H3K27me3. It is also crucial in the progression of diseases in humans. Pathological progression of cancer is characterized by aberrant histone methylation in cancer cells, and dysregulated activities of histone methyltransferases are often linked to carcinogenic transformation. Silencing of antiviral genes is caused by the loss of H3K36me2, accompanied by a counterbalancing increase in H3K27me3. Valemetostat or pharmacological inhibition of the enhancer of zeste homologs EZH1/2 reinstates tumor suppressor gene functionality by inhibiting the repression of H3K27me3.
-
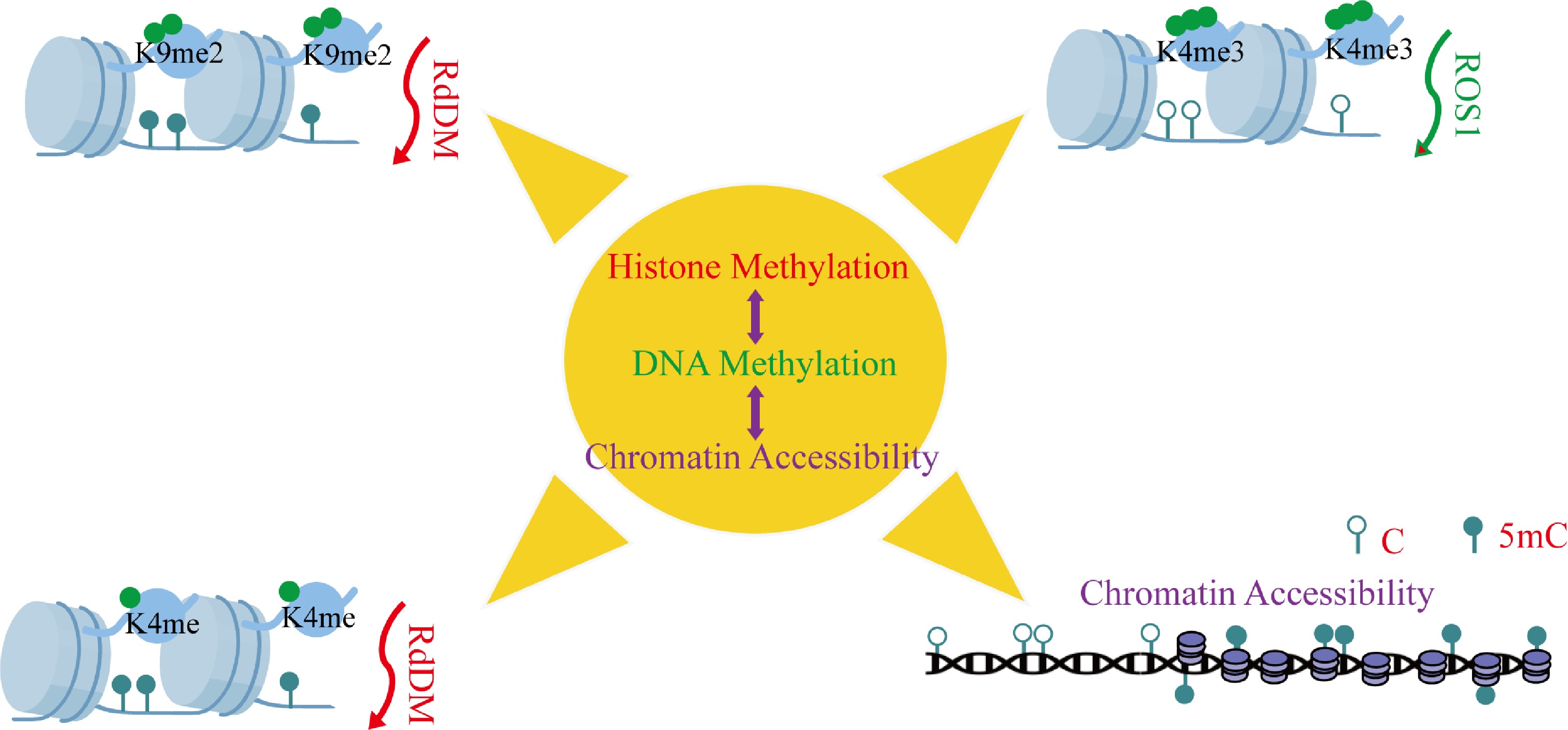
Figure 3.
Synergistic regulation between DNA methylation and histone modifications. This depicts the synergetic control between histone modifications and DNA methylation. These essential interactions are: (i) RdDM-mediated small RNA-dependent de novo DNA methylation, (ii) H3K9me2 deposition and H3K4me establishment, (iii) H3K4me3-mediated hiring of DNA demethylases, and (iv) enrichment of histone modifications by DNA methylation. Abbreviations: 5mC, 5-methylcytosine; C, cytosine.
-
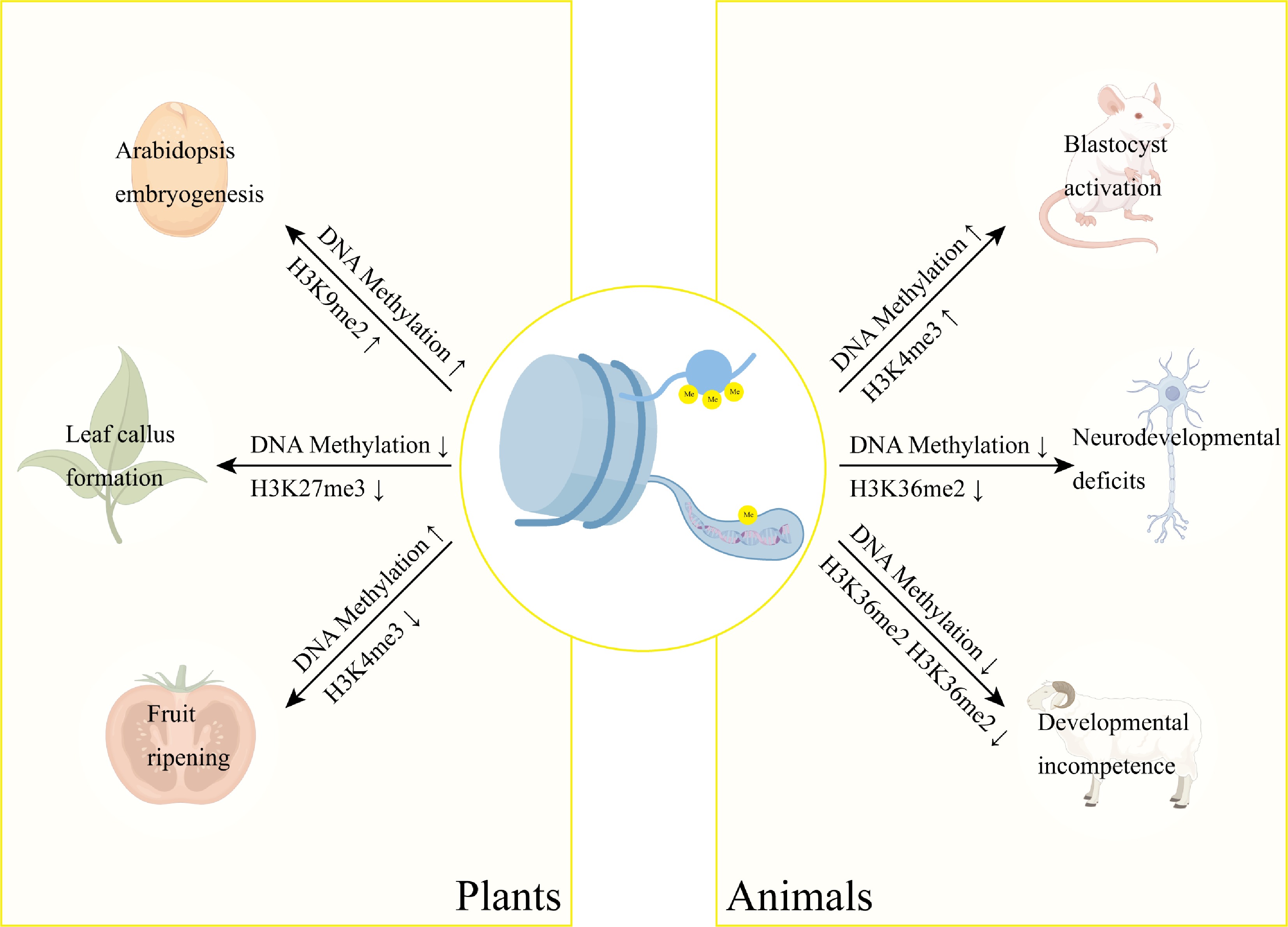
Figure 4.
Interplay between DNA methylation and histone methylation in developmental regulation. DNA methylation and histone methylation function synergistically in gene regulation. These epigenetic modifications form an integrated regulatory network controlling key developmental processes across species, including Arabidopsis embryogenesis, peach somatic embryogenesis, and tomato fruit ripening. Notably, their coordinated action is equally essential in mammalian systems, regulating mouse oocyte activation, neural development, and somatic cell reprogramming in cloned goats.
Figures
(4)
Tables
(0)