-
Dandelion is a perennial herb of compositae family, native to Europe and widely grown in temperate regions of the northern hemisphere[1]. In Chinese traditional medicine books, its effects of dispelling wind-heat, detoxification and detumescence, diuresis, defecation, jaundice, liver and gallbladder detoxification ability are comprehensively described in detail, and as a medicinal plant, it has anti-inflammatory, anti-oxidation, anti-tumor, antibacterial, hypoglycemic, liver and gallbladder, regulation of gastrointestinal microecology and other therapeutic effects[2−4], so it has been widely investigated as an ingredient in the health care industry for various foods.
Dandelion is rich in a variety of bioactive substances, including polyphenols (trans-p-hydroxyphenylpropofol, trans-p-hydroxyphenylacrolein, p-hydroxybenzoic acid, p-hydroxyphenylpropionic acid, protocatechualdehyde), flavonoids (rutin and quercetin), polysaccharides and triterpenoids, etc. Moreover, the types and content distribution of functional components in different tissues are different[5], leading to different roles. According to the existing research results, dandelion leaves are rich in caffeic acid, coumarin, chlorogenic acid, flavonoid glycosides, and chicoric acid; dandelion roots contain triterpenoids, chlorogenic acid, caffeic acid, rutin, and vanillic acid; dandelion flowers are rich in flavonoids, free luteolin, chelonethylene glycol, and chrysol[6−8]. It is widely believed that dandelion leaves can be used as a gallbladder, diuretic, and bitter digestive irritant, while the root is also used as an alternative to help relieve skin diseases, treat digestive disorders, increase bile flow, and can promote appetite[9,10]. Considering the differences in the types and distribution of compounds, a more effective way of dandelion utilization is to divide the whole plant into different parts for individual bioactivity compound extraction and specific beneficial effects evaluation. The results of compound identification and differentiation in dandelion tissue using different spectral techniques and multivariate statistical techniques have not yet been systematically studied.
Spectroscopic techniques used for compound identification mainly include infrared spectroscopy (different chemical functional groups absorb different frequencies of infrared light, the optical technology can detect the vibration and rotation of molecular bonds, and can be used for chemical fingerprint identification, chemical imaging and chemical structure analysis) and fluorescence spectrum (each organic compounds has a separate maximum emission / excitation wavelength at different wavelengths, parallel factor analysis (PARAFAC) was used to process the excitation-emission matrices (EEMs) and determine the characteristics of samples)[11].
Fourier transform infrared spectroscopy (FTIR) is a vibration spectroscopy technology based on the mathematical processing of Fourier transform, which has been widely used in food quality control, food structure and function research due to its characteristics of high speed, high accuracy and resolution. Studies have shown that FTIR spectrum combined with a variety of analysis methods, such as principal component analysis (PCA, a statistical technique to reduce the dimension of data, can use less dimensions to describe the change of data, but still contains most information[12]) and partial least squares regression (PLS-R, a multiple linear regression analysis, reduce the raw data to less and maximizing the explained variance to detect the relationship between predictor and response variables[13]). It can accurately and rapidly predict edible oil adulteration, distinguish coffee bean samples from different origins, detect meat and meat product adulteration, monitor biochemical, microbial spoilage and shelf life, and determine protein and lipid structure changes[14−17].
Three-dimensional fluorescence spectrum (known as excitation-emission matrix) could quickly determine the fluorescence intensity of a substance in different excitation wavelength and emission wavelength ranges, and mixtures can be directly detected and distinguished by the different fluorophores structures of each compound and the corresponding three-dimensional fluorescence spectral information[18]. Parallel factor analysis is an efficient method to decompose fluorescence excitation-emission matrices into their underlying chemical components, this analysis method can lead to the mathematical identification and quantification of independently varying fluorophores (individual component with fluorescent group) from the complex, obtain excitation and emission spectra and their corresponding concentration or content[19]. Three-dimensional fluorescence spectroscopy combined with parallel factor analysis has the advantages of high sensitivity, good selection performance and no damage to samples, and has been widely used in the detection and identification of chemical components in vinegar, wine and other foods as well as water quality assessment[20−22].
Without sample pretreatment and chromatographic conditions optimization, or time-consuming mass spectrum processing and compound identification, the Fourier transform infrared spectroscopy and three-dimensional fluorescence spectrum combined with corresponding analytical methods have been widely used in the extraction and classification of plant species, tissues and functional compounds. The spectroscopy technologies have been used by thousands of researchers for commercial or academic purposes, but their application scope and analysis results have not yet been systematically analyzed, which also limits the mining and comparison of spectral data and hinders the correct technical selection. In this paper, dandelion roots, stems, flowers and leaves were obtained and prepared for water extraction preparation, the FTIR spectroscopy combined with PCA analysis, and three-dimensional fluorescence spectroscopy combined with PARAFAC analysis were used to establish corresponding models to identify the extracts of dandelion tissues, so as to compare the classification results of different tissues (roots, stems, flowers and leaves) of dandelion. Based on the research of substance species and content distribution in dandelion tissues, this paper tried to fill the gap between molecular spectroscopy discrimination results and substance differences, without comparing individual compound contents in each tissue. In addition, the discrimination capacity difference between Fourier transform infrared spectroscopy and three-dimensional fluorescence spectrum were discussed, and the internal relationships between molecular bond vibrations and rotations spectrum and fluorescent compounds structure and content contour were also investigated. In summary, this paper aims to compare the Fourier transform infrared spectroscopy and fluorescence spectroscopy analysis results systematically.
-
Dried dandelion plant was purchased from a local pharmacy, and stored in sealed light resistant packaging at 4 °C before use. Formic acid, deuterium oxide for infrared spectrum and three-dimensional fluorescence spectrum were of chromatographic grade and obtained from Shanghai Maclin Biochemical Co., Ltd. (Shanghai, China). Deionized water used to prepare solutions was 18 MΩ and purified using an ultra-pure water system.
Preparation of dandelion tissue extracts
-
Different dandelion tissue extracts were prepared according to previous research literature[23]. The roots, stems, flowers and leaves of the whole dried dandelion were cut out and oven-dried separately at 60 °C until the dry weight was constant. The same tissues from different plants were milled, and the resulting particles were mixed and screened using a 60-mesh sieve. Six grams of root, stem, flower and leaf powder was mixed with 60 mL of 1‰ formic acid water solution separately, and vortexed at 20 °C for 2 h. After centrifugation at 8,000 r/min for 15 min, the supernatants of different dandelion tissues were taken and filtered through a 0.22 μm nylon filter to yield the crude extract, and all the processed extracts were stored at 4 °C in the dark.
Fourier transform infrared spectroscopy acquisition
-
The obtained tissue extracts were freeze-dried, 0.4 g of the samples were dissolved in 5 mL of deuterium oxide, then filtered with a needle filter of 0.22 μm before testing. About 3 mL of liquid was placed in a diamond ATR module, and the FTIR spectrum was collected by Bruker Tensor 27 Fourier Infrared spectrometer. Instrument parameters were set as follows: the wavelength range was 4,000−400 cm−1, with a spectrum resolution of 1 cm−1, the scan number was set as 32, the temperature was 25 °C, and the spectrum of deuterium oxide was used as the blank sample[15, 16].
Principal component analysis
-
The principal component analysis was performed in the window between 3,900 and 400 cm−1. Baseline were first corrected for all samples, then the spectra were normalized (all spectra were centered on the mean, and the mean absorbance was calculated and then subtracted from the spectrum. The FTIR spectra were scaled to make the sum squared deviation over the indicated wavelengths equal one.) and smoothed with polynomial 2nd order using the Savitzky-Golay-algorithm[24]. Then the covariance matrix of normalized spectra was computed to identify the variables with respect to others. The eigenvectors and eigenvalues of the covariance matrix were calculated and ordered by eigenvectors values in descending order, to determine the proper principal components in order of significance. Recast the data along the axes of principal components using the eigenvectors of the covariance matrix, which could be done by multiplying the transpose of the original data set[25].
PCA was used to decompose the data matrix, and covariance data matrices were used to calculate the principal components (PCs). The principal components of PC1 to PC3, and their corresponding PC loadings were calculated. The PCA analysis was conveyed by the additional automatic application following the above steps, and the results were illustrated using the software of Origin 9.0 (Origin Lab, Northampton, USA).
Three-dimensional fluorescence spectrum acquisition
-
The processed extracts of root, stem, flower and leaf obtained from whole dried dandelion were diluted 20 times to obtain the tested liquid for three-dimensional fluorescence spectrum measurement. The fluorescence analysis was performed using the PerkinElmerLS55 system with 1 cm quartz colorimeter, the device was validated with deionized water, the Raman and Rayleigh peaks were measured and used to correct the original spectrum. Then the tested extracts of different tissues were placed at the excitation wavelength of 200−400 nm and the emission wavelength of 220−600 nm to obtain the three-dimensional fluorescence spectrum. The excitation interval was set at 10 nm, the voltage was set as 700 V, and the excitation and emission slit width were both 20 nm.
Parallel factor analysis
-
Parallel factor analysis statistically decomposes the three-dimensional fluorescence spectrum into individual fluorescence components and a residual matrix. The individual fluorescence components are directly proportional to the component concentration in the sample and could be converted into actual concentration when the excitation and emission of each component are known[19, 26].
$ {\mathrm{x}}_{\mathrm{i}\mathrm{j}\mathrm{k}}=\sum _{\mathrm{n}=1}^{\mathrm{F}}{\mathrm{a}}_{\mathrm{i}\mathrm{n}}{\mathrm{b}}_{\mathrm{j}\mathrm{n}}{\mathrm{c}}_{\mathrm{k}\mathrm{n}}+{\mathrm{\varepsilon }}_{\mathrm{i}\mathrm{j}\mathrm{k}} $ where, xijk is the fluorescence intensity of the ith dandelion extraction at the kth excitation and jth emission wavelength, ain is directly proportional to the concentration of the nth fluorophore in the ith sample, bjn and ckn are estimates of emission and excitation spectra of nth fluorophore at wavelength j and k. F is the number of components, and εijk is the residual matrix.
In this paper, the PARAFAC analysis was conveyed using the N-way program[26] following the steps of blank spectrum subtracting, outliers removed, limiting the Raman scattering, data normalization, and parallel factor analysis model establishing using the inner automatic functions. After subtracting deuterium oxide spectrum manually, and loading the total three-dimensional fluorescence spectrum containing 379 × 21 intensity readings, the program first eliminated Rayleigh and Raman scattering peaks of each scan centered on the respective scattering peak by excising portions (10 and 20 nm at each excitation wavelength). The PARAFAC model was then established with default PARAFAC constraints, no negative values in concentration, emission and excitation wavelength were applied to process the data. The PARAFAC model was tested from one to five component by means of fitting values, core consistency, and split-half quality calculation. Samples with high leverage (the elements on the diagonal of the hat matrix of the score matrix) or high sum-squared residual were removed until no samples were assessed as outliers by default set, and the PARAFAC model of proper component number was identified.
Statistical analysis
-
All the tissue extracts of dandelion root, stem, flower and leaf for FTIR, and three-dimensional fluorescence spectrum acquisition were repeated three times, and the data were expressed as mean ± standard deviation. The PCA analysis was carried out using the additional application in the software of Origin 9.0, and the PARAFAC analysis was conveyed by the N-ways program.
-
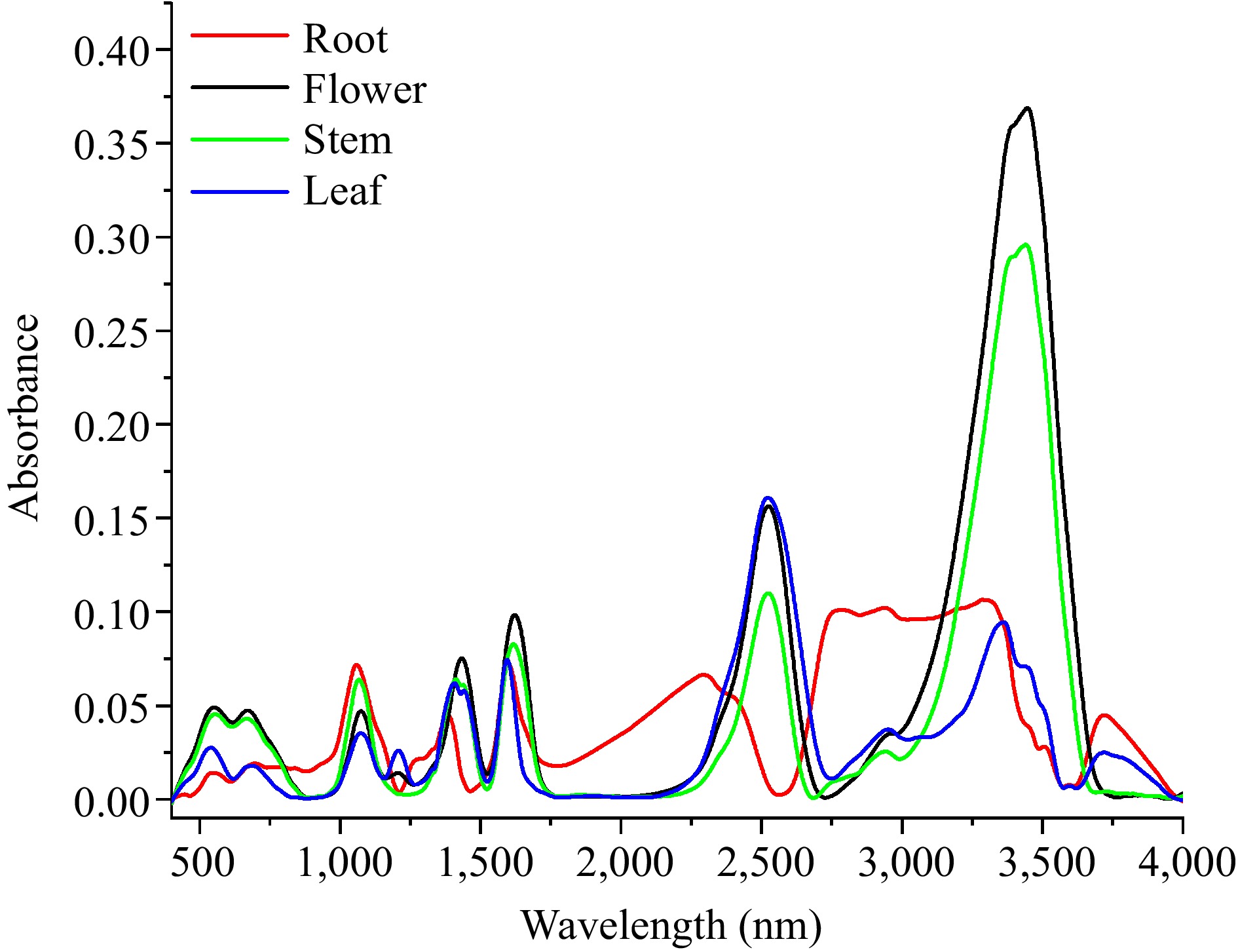
The Fourier transform infrared (FTIR) spectroscopy of dandelion tissues extracts of root, stem, flower and leaf were measured respectively, and their absorbance spectrum was recorded. As shown in Fig. 1, the FTIR of dandelion stem, flower and leaf have the same peak location wavelength and similar fingerprint, the dandelion root had two distinct peak wavelengths between 2,500−1,750 cm−1 and 3,500−2,750 cm−1. The absorbance spectrum indicated that stem, flower and leaf might have the same compounds with different content, while the substances in root extracts were different.
Since the differences in absorbance spectra are due to compound species and contents of compounds in different tissues, the identification efficiency was evaluated by principal component analysis (PCA). As shown in Fig. 2, two main components of PC1 (factor 1, 59%) and PC2 (factor 2, 38%) were extracted according to the above steps in the method section with the cumulative variance contribution rate of 97%, which indicated that the two-component model could explain the total 97% of the absorbance spectroscopy difference determined by the dandelion tissues, and could be used to distinguish the test dandelion tissues. Although obvious difference existed between the three repeated experiments of stem, flower and leaf tissue extracts, the PCA maps showed the Fourier transform infrared spectroscopy characteristics of dandelion tissues. Dandelion leaf, represented by green points, accounted for the comparatively higher values in PC1 and PC2 dimensions, ranging from 0.5 to 0.8 and −0.2 to 0 respectively; the dandelion stems (marked as blue points) values varied in the moderate range, which changed in the range of −0.1 to 0.3 in the PC1 axis and −0.4 to −0.1 in the PC2 axis. For the dandelion flowers in the red group, its values were of the comparatively lower range, showing a narrow area in the PC1 coordinate (−0.2 to 0) and a large area in the PC2 coordinate (−0.3 to −0.6). The dandelion root (black points) group showed a different tendency compared to the other tissues, with a smaller repeat difference and high values in both PC1 and PC2. According to the PCA analysis results, dandelion tissue extract could be divided into three groups, dandelion root group and dandelion leaf were not well disguised from each other.
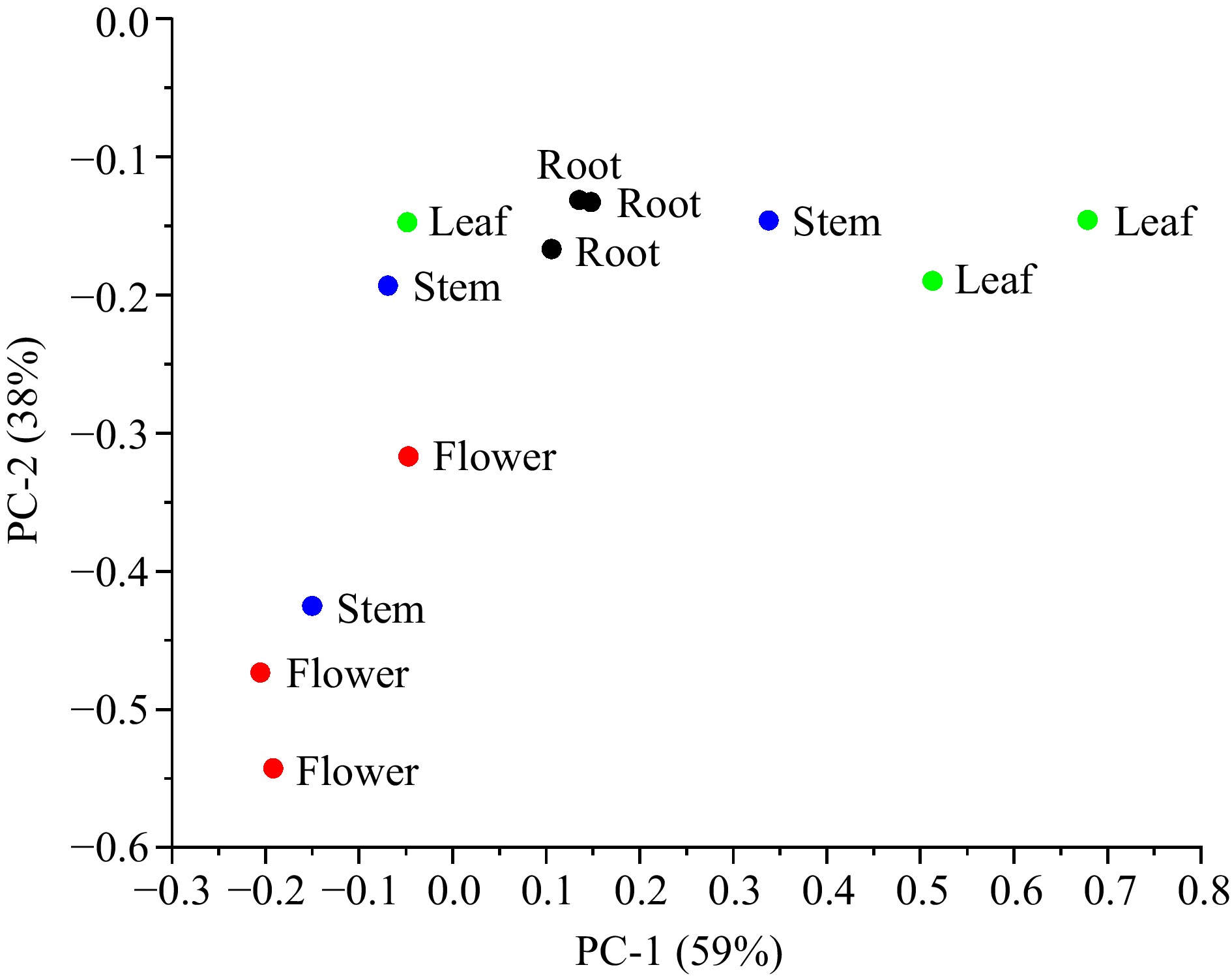
Figure 2.
Score cluster plot with top two principal components (PCs) for different dandelion tissues.
Fluorescence spectrum measurement
-
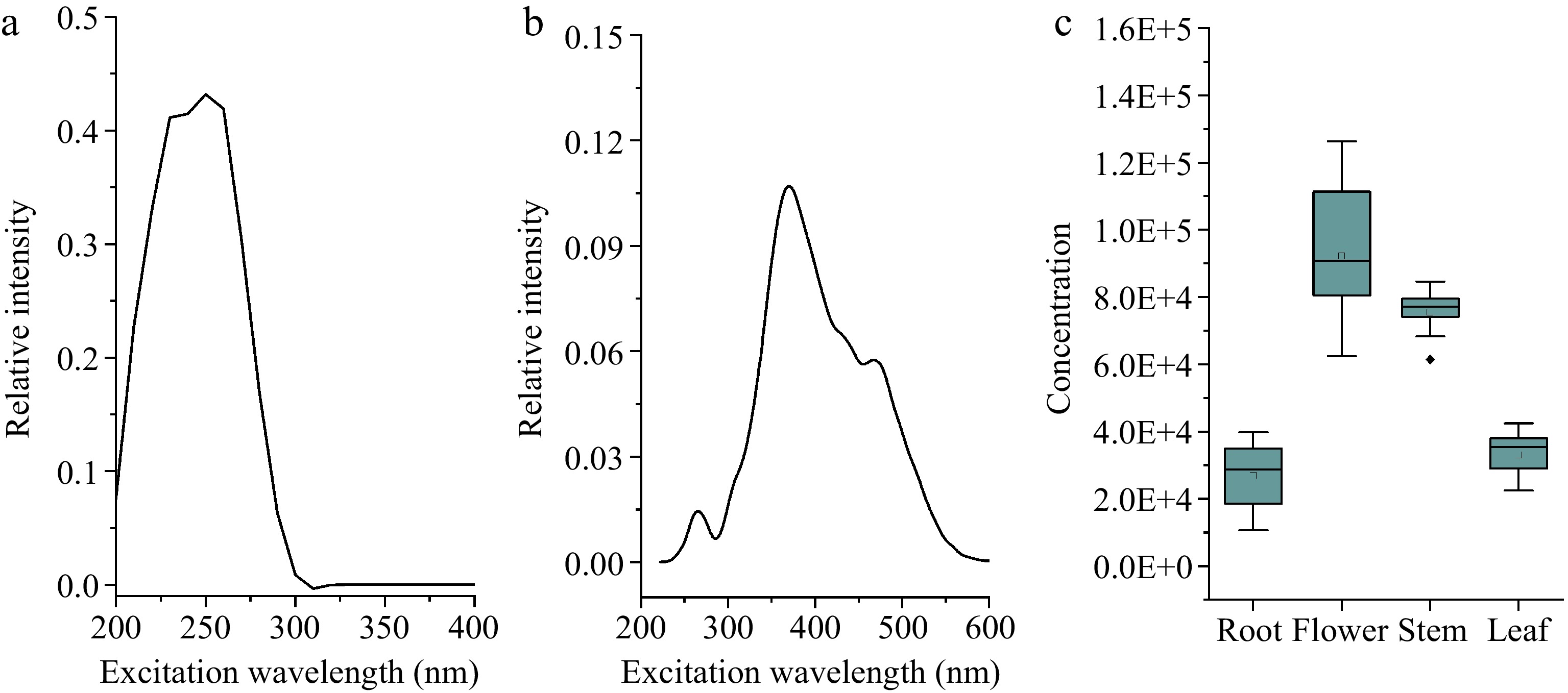
Different dandelion tissue extracts were scanned to obtain raw three-dimensional fluorescence spectra (excitation-emission matrices, EEMs) in the excitation range of 200−400 nm and the emission range of 220−600 nm. As shown in Fig. 3, where a, b, c, and d correspond to the spectrum of dandelion root, flower, stem, and leaf extracts respectively. The fluorescence spectra of dandelion tissue extracts showed different fluorescence fingerprints with one or two relatively intense bands, the fluorescence spectra of dandelion root and leaf had one strong band and maximum excitation / emission wavelengths of about 260 nm / 370 nm with obvious intensity difference, while the dandelion stem and flower fluorescence spectrum had two intense bands at 220 nm / 370 nm and 260 nm / 370 nm, and the excitation wavelength at 220 nm had higher fluorescence intensity compared to the ones at 260 nm. In addition, the fluorescence intensity of all the tissues had a fluorescent band around 260 nm / 370 nm, and the obtained three-dimensional fluorescence spectrum could be divided into two distinct groups with different intensity bond distribution and fluorescence intensity.

Figure 3.
Three-dimensional fluorescence spectra of dandelion tissue extracts. (a) Root, (b) flower, (c) stem, (d) leaf).
PARAFAC analysis
-
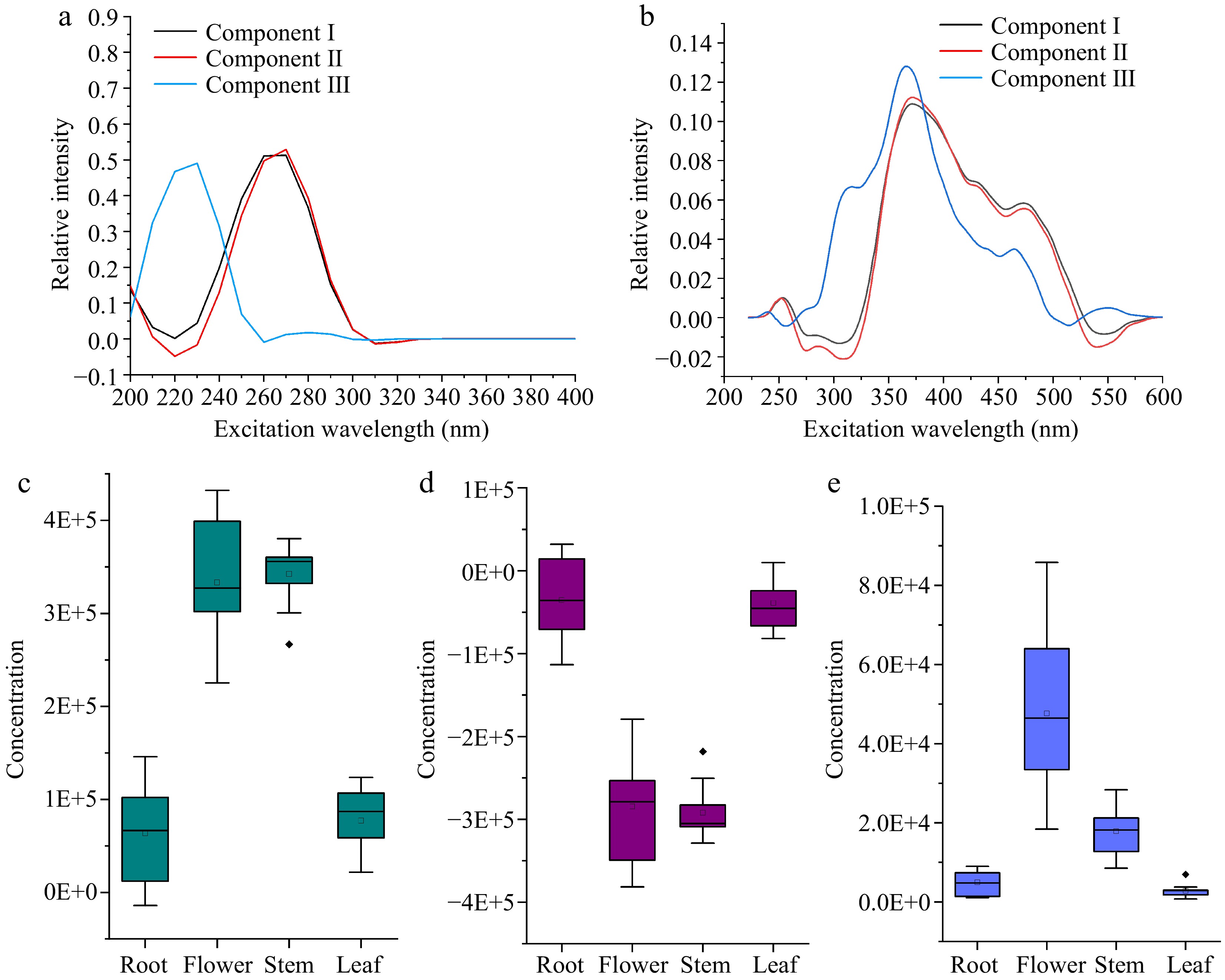
The PARAFAC model was established from component number 1-5 after removing Rayleigh and Raman scattering from the original fluorescence spectra. In order to determine appropriate component numbers, the residual sum of the square, core consistence and interaction number were compared and evaluated. As shown in Fig. 4, the residual sum of the squares decreased as more components were selected. The core consistency analysis of the model reaches 100 at the first component and decreased in the second, third, fourth, and fifth component models, while the interaction number remained at the lowest level at one- component model, followed by the second and third component models. Considering all the test indicators, one and three component models were identified for the fluorescence dataset based on the higher residual sum of the square and lower interaction number. The core consistence retained some variability, while the four and five component models were rejected due to their lower residual sum of square and higher interaction number. Although one and three components were selected to model, it does not indicate that only one or three types of fluorophores were present in these extracts[27].
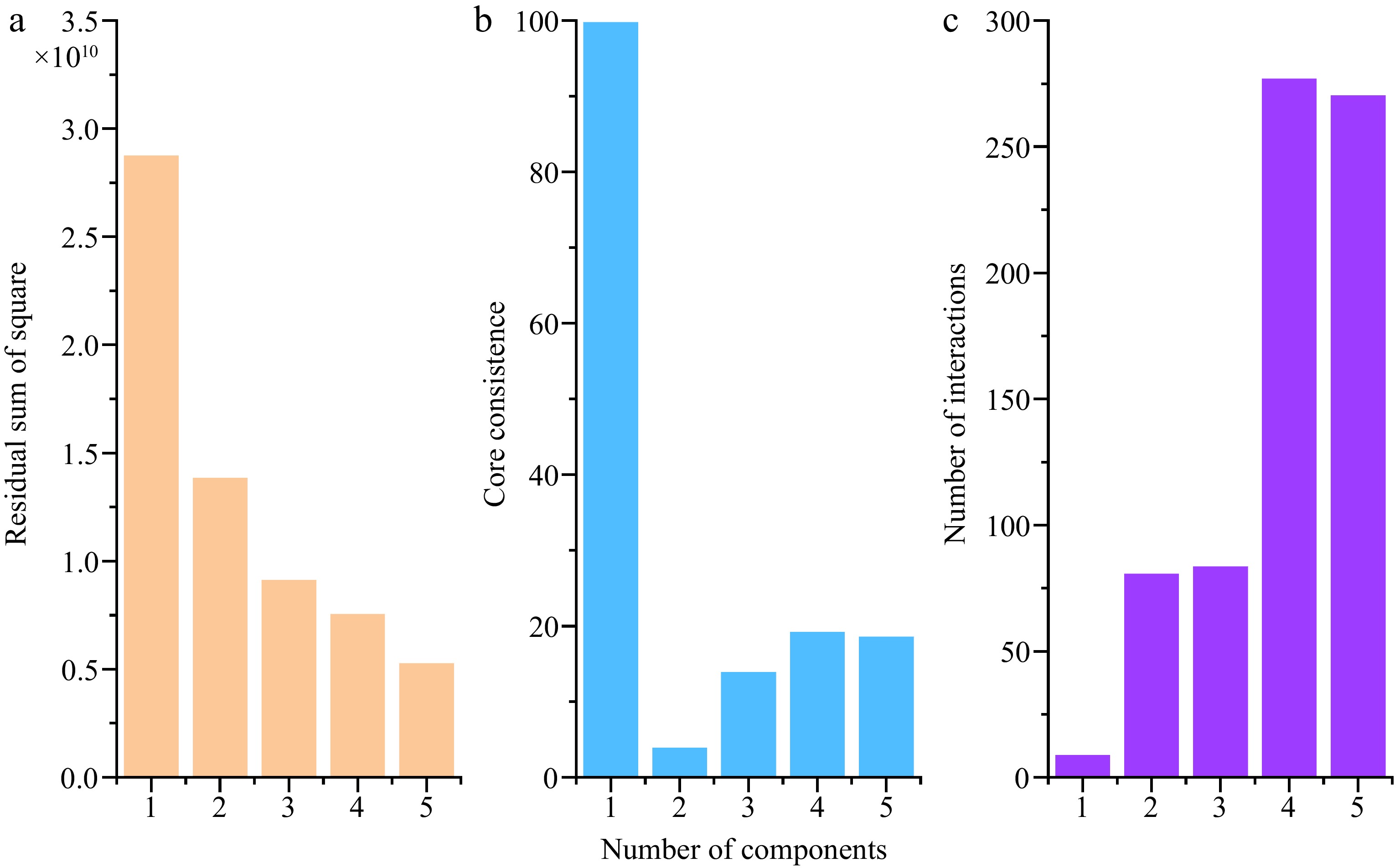
Figure 4.
Analysis diagram of the parallel factor model. (a) Square residual, (b) core consistence, (c) interaction number.
Figure 5 showed the excitation and emission spectrum and calculated concentrations determined by the one component PARAFAC model. The maximum excitation wavelength of the first component was about 260 nm, and the maximum emission wavelength was about 370 nm, and the obtained concentration of dandelion flower extract was the highest, followed by tissues of stem, leaf and root. In addition, the obtained concentration of dandelion flower and stem, root and leaf overlapped with each other, this result indicated that one-component model cannot distinguish tissues of flower and leaf tissue, as well as root and leaf. Then the three-component model was established and shown in Fig. 6. The first component represented by the black line had a maximum excitation wavelength at about 260 nm and a maximum emission wavelength at about 370 nm, which were the same as the one component model, the second component marked with the red line had a maximum excitation wavelength at about 270 nm and a maximum emission wavelength at about 370 nm, while the third component of the blue line had a maximum excitation wavelength at about 230 nm and a maximum emission wavelength at about 350 nm. The concentration distribution was the same as the component one model, for the stem tissue had the highest concentration, followed by tissues of flower, leaf, and root respectively. While the second component had the opposite tendency, the concentrations of root and leaf were higher compared to the flower and stem, and the concentration distribution of the third component was the same as the first, where the flower and stem concentration had a higher value, than root and leaf.
-
Fourier transform infrared spectroscopy with principal component analysis, and three-dimensional fluorescence coupled with parallel factor analysis have been used to identify plant species and origin or determine the effects of processing on food quality. In this paper, these two methods were used to process spectrum data, and evaluate the component characteristics of different dandelion tissues.
Fourier transform infrared spectroscopy coupled with PCA analysis could well distinguish the stem and flower, but failed to discriminate the root and leaf tissue. Former research proved that dandelion root, leaf, and flower contain polyphenols and flavonoids, while dandelion root contains unique triterpenoids, and dandelion leaf contains unique flavonoid glycosides[5]. The peak list contains O-H aromatic (3,400 cm−1), C-H aromatic (2,900 cm−1), C-H aliphatic (2,800 cm−1), C=O (1,743 cm−1), C=C (1,640 cm−1), C=C aromatic (1,550 cm−1), C-O (1,100 cm−1), C-H alkanes (1,450 cm−1) and C-N (1,240 cm−1), these functional groups indicated that phenolic acids, alcohols, esters, carboxylic acids widely exist in the extracts of various dandelion tissues, which is also proven by former research on FTIR analysis results[28]. The FTIR spectrum includes absorption, reflection, emission, or photoacoustic spectrum, and all the substance species and content difference contribute to the spectrum characteristics. On the other hand, PCA summarises the obtained data features, without reference to prior knowledge about whether the samples come from the same dandelion tissues, or the species and content of polyphenol flavonoids, triterpenoids and lavonoid glycosides[29]. Therefore, the dandelion origin, experiment error, and even repeat number could all impact the PCA analysis results. As shown in Fig. 2, two dimensional principal component analysis (PCA) was used to analyze the difference of extracts from different tissues of dandelion. The obtained information is the trend of the point pattern relative to the other patterns, the close point distance of tissue sample reflects the higher similarity among tissues, while the far sample distance of each point represents a comparatively obvious difference[12]. In this case, it is a combination of similarity and difference in FTIR spectral patterns. The higher repeat difference might be related to the lower classical PCA efficiency, it implies that the PCA model failed to classify different tissues. Increasing the number of repeated experiments could improve the discriminant efficiency and the robustness of the model, but the calculated principal component values and the distribution in the score cluster graph will not change.
Three-dimensional fluorescence spectrum showed all the fluorescent compound fingerprints at specific excitation and emission wavelengths, with various bond distribution and fluorescence intensity. Based on the available fluorescence data and former research, it could be referred that polyphenols show fluorescent peaks at 220 nm / 370 nm, with a higher maximum intensity in flower extracts, than the ones of stem, leaf and root[23, 30]. Although other compounds of flavonoids, triterpenoids and lavonoid glycosides have unique fluorescence absorption and emission, their maximum wavelength of excitation and emission are closed to typical polyphenol, and cannot be detected or discriminated by the fluorescence scan[31].
In order to compare and distinguish the fluorescence spectra of different dandelion tissues, the PARAFAC method was then used to extract the characteristic components and corresponding spectral features. When compared to the PARAFAC models, the obtained concentrations of leaf and root, as well as the stem and flower tissues overlapped with each other, in both one- (Fig. 5c) and three- (Fig. 6c−e) component models. This result indicated that calculated one- and three-component models had similar efficiency, since we cannot distinguish dandelion tissues in box plot of one component or all three components. On the other hand, the calculated excitation and emission spectra of the first component in the one-component model were the same as the third component in the three-component model, and both fluorescence spectra referred to the existence of classical phenolic compounds. The fluorescence spectrum only reflects the fluorescent component characteristics, this why the PARAFAC method has only successfully determined dissolved organic matter and fingerprint in wastewater or water processing[32].
Based on the results and discussion, it seems that the FTIR-PCA analysis had a better discrimination capacity, since all the substances including fluorescent components and non-fluorescent components could be taken into account, while the fluorescence spectrum does not have enough resolution for discrimination substances with similar structures with close excitation and emission peak wavelengths. By systematically comparing the above methods, this paper gives the limitation and potential application in plant tissue discrimination, as well as the processing effects, plant origin, and compound evaluation in the food industry.
-
Fourier transform infrared spectroscopy combined with principal component analysis, and three-dimensional fluorescence spectroscopy combined with parallel factor analysis were used to distinguish different dandelion tissue extracts. Results indicated that FTIR-PCA analysis could well discriminate tissues of stem and flower from leaf and root, while three-dimensional fluorescence spectrometry with PARAFAC analysis cannot differentiate the tissues of leaf from root, and stem from flower, for the concentrations overlapping with each other. This paper demonstrates that both methods could distinguish samples without prior knowledge of the substance type and content, and FTIR-PCA might be more suitable when fluorescent components exist in various amounts among different samples.
-
The authors confirm contribution to the paper as follows: study conception and design: Li G, Zou H; data collection: Li G; analysis and interpretation of results: Li G, Zou H; draft manuscript preparation: Li G, Chen Y. All authors reviewed the results and approved the final version of the manuscript.
-
All data generated or analyzed during this study are included in this published article.
This work was supported by funding: 'Innovation Project of Shandong Province Agricultural Application Technology', No 2130106.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press on behalf of China Agricultural University, Zhejiang University and Shenyang Agricultural University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Li G, Zou H, Chen Y. 2023. Discrimination capacity analysis of FTIR-PCA and EEM-PARAFAC on dandelion tissues extracts. Food Innovation and Advances 2(4):247−254 doi: 10.48130/FIA-2023-0026
Discrimination capacity analysis of FTIR-PCA and EEM-PARAFAC on dandelion tissues extracts
- Received: 19 May 2023
- Accepted: 08 September 2023
- Published online: 25 October 2023
Abstract: Dandelion root contains triterpenoids, polyphenols and flavonoids, dandelion leaf is rich in polyphenols, flavonoids, flavonoids glycosides, and dandelion flower mainly contains flavonoids, among other substances. These different substance content leads to specific benefits and function effects of each part. Fourier transform infrared spectroscopy, three-dimensional fluorescence spectroscopy and related multivariate statistical methods are widely used to determine sample characteristics, but limited research focuses on the substance difference and characteristics in dandelion tissues. In this paper, Fourier transform infrared spectra-principal component analysis and three-dimensional fluorescence spectroscopy-parallel factor analysis were conveyed to analyze dandelion stem, leaf, root and flower tissue extracts, for determining the substance species and content difference among dandelion tissues and evaluating the discrimination capacity of these analysis methods. The Fourier transform infrared spectroscopy of root was distinct from others, and the two principal component models could distinguish dandelion stem and flower, but failed to differentiate leaf and root; while the excitation and emission matrix showed that stem and flower, leaf and root had similar intensity band distribution but different fluorescence intensity, and the parallel factor analysis results proved that one- and three-component models cannot differentiate the tissues of stem and flower, leaf and root, since the fluorescent compounds (polyphenol, flavonoid etc.) structure and content were similar in different tissues. These results indicated that Fourier transform infrared-principal component analysis might be a useful method when various fluorescent compounds exist.