-
Microascales was introduced by Benny & Kimbrough[1]. Currently, seven families, Ceratocystidaceae, Chadefaudiellaceae, Gondwanamycetaceae, Graphiaceae, Halosphaeriaceae, Microascaceae, and Triadelphiaceae are accepted in the order[2−5].
Graphiaceae was previously published by de Beer et al.[6] with nine accepted Graphium species. However, the family is currently recorded as 'Nom. inval., Art. F.5.1 (Shenzhen)' with Editorial comment as 'An identifier issued by a recognized repository was not cited in the protologue'' based on Index Fungorum[7].
Triadelphiaceae was introduced to accommodate a monotypic genus Triadelphia[8]. The family is characterized by hyphomycetous, holoblastic, monoblastic, fusiform to cylindrical or clavate, hyaline to pale brown conidiogenous cells, acrogenous with one to five different forms of conidia[8−10]. Chuaseeharonnachai et al.[2] introduced Triadelphia hexaformispora, a new species from a freshwater habitat in northern Thailand and a new genus Synnematotriadelphia, to accommodate two synnematous Triadelphia species, T. stilboidea and T. synnematofera under the new genus name, based on both morphology and molecular data.
Currently, 20 epithets are listed in Triadelphia[7], being mostly reported from terrestrial habitats. Triadelphia fusiformis, T. heterospora, T. hexaformispora and T. morgoensis have been reported from freshwater habitats[2,8]. In this study, a new Triadelphia species was collected from terrestrial decaying wood and is introduced based on both morphological characteristics and multi-gene analyses. Morphological descriptions and illustrations are provided. The current taxonomic placement of Microascales is also clarified. In addition, a new family, Synnematotriadelphiaceae is introduced and Graphiaceae is validated herein.
-
Fresh samples were collected from decaying wood in a terrestrial habitat in Mukdahan Province, Thailand. The samples were processed following the methodology as described by Boonmee et al.[11]. A Carl Zeiss GmbH stereo microscope (SteREO Discovery. V12, Germany) fitted with an AxioCam 105 color camera was used for image capture. Fungal micromorphology was examined using a Leica EZ4 Series stereomicroscope and photographed with a Nikon ECLIPSE 80i compound microscope (Nikon, Japan) fitted with a NikonDS-Ri2 digital camera (Nikon, Japan). Measurements of microscopic characters were calculated using the Tarosoft (R) Image Frame Work (IFW) software version 0.97 and a photoplate was made using Adobe Photoshop CS6 version 13.1.2. software (Adobe Systems, USA).
Single spore isolations were obtained following the methodology as described by Senanayake et al.[12]. Germinating spores were aseptically transferred to fresh malt extract agar plates (MEA: 33.6 g/l sterile distilled water, Difco malt extract) and incubated at 25 °C. Cultures were incubated for 28 d and morphological characters, such as colour, colony shape, and texture were recorded. Type materials were deposited in the Herbarium of Mae Fah Luang University (Herb. MFLU), Chiang Rai, Thailand and Ex-type living cultures were deposited at Mae Fah Luang University Culture Collection (MFLUCC). Faces of fungi and Index Fungorum numbers were obtained as described in Jayasiri et al.[13] and Index Fungorum[7].
DNA extraction, PCR amplification and sequencing
-
Genomic DNA was extracted from fungal mycelium using PureLink™ Quick Gel Extraction and PCR Purification Kit (Invitrogen by Thermo Fisher Scientific) following the manufacturer's instructions. Five gene regions were amplified with primer pairs as following: the internal transcribed spacer (ITS) region with primer pairs ITS5 and ITS4[14], the large subunit nuclear ribosomal DNA (LSU) with primer pairs LROR and LR5[15], the small subunit ribosomal DNA (SSU) with primer pairs NS1 and NS4[14] and the partial RNA polymerase II subunit (RPB2) with primer pairs fRPB2-5F and fRPB2-7cR[16].
The polymerase chain reaction (PCR) thermal cycle program for ITS, LSU and SSU amplification were initially 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 50 s, elongation at 72 °C for 1.30 min and final extension at 72 °C for 10 min. The PCR thermal cycle program for RPB2 was initially 95 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 1 min, annealing at 52 °C for 1 min, elongation at 72 °C for 1.30 min and final extension 72 °C for 10 min. The quality of amplified PCR products was checked on 0.85% agarose gel electrophoresis. The PCR products were sent for sequencing at Macrogen Inc. (Geumcheon-gu, Seoul, Korea).
Phylogenetic analysis
-
Phylogenetic analysis of the combined aligned dataset of ITS, LSU, SSU, and RPB2 sequence data was analyzed based on maximum likelihood (ML) and Bayesian inference (BI) following the methodology as described by Mapook et al.[17]. The sequence data of closest relative strains were selected and downloaded from GenBank based on the latest treatment and updated accounts of Microascales with updated accounts of the genus Triadelphia from recent relevant literature[2,8,10]. The sequences used for analyses with GenBank accession numbers are given in Table 1. Phylogenetic trees were drawn using FigTree 1.4.2[18] and edited by Microsoft Office PowerPoint 2019 and Adobe Photoshop CS6 (Adobe Systems, USA). The new nucleotide sequence data is deposited in GenBank.
Table 1. Taxa used in this study and their GenBank accession numbers. New sequences are in bold.
Taxa Strain no.1 GenBank accession numbers2 ITS LSU SSU RPB2 Acaulium albonigrescens CBS 109.69 KY852469 KY852480 − − Ambrosiella xylebori CBS 110.61 − EU984294 AY858659 − Ceratocystis fagacearum CMW 2656 − KM495341 − − Corollospora marina AFTOL-ID 5008 − AY762985 FJ176843 − Corollospora ramulosa PP1232 − AF491276 − − Falcocladium multivesiculatum CBS 120386T EU040217 JF831932 JF831928 − Falcocladium sphaeropedunculatum CBS 111292T JF831938 JF831933 JF831929 − Faurelina elongata CBS 126.78 GU291802 DQ368625 DQ368657 DQ368639 Faurelina fimigena CBS 352.78T MH861149 − − − Graphium fimbriasporum CMW 5605T NR_155108 NG_067529 NG_064882 − Graphium fimbriisporum CMW 5606 AY148180 − AY148172 − Graphium laricis CMW 5601T AY148183 NG_064370 AY148162 − Graphium penicillioides CBS 102632T KY852474 NG_067324 − − Graphium pseudormiticum CMW 503T NR_136962 NG_064371 NG_064881 − Knoxdaviesia capensis AFTOL-ID 1907 − FJ176888 FJ176834 − Knoxdaviesia proteae CBS 486.88T AY372072 AF221011 AY271804 − Knoxdaviesia serotecta CBS 129738T MH865536 MH876970 − − Knoxdaviesia ubusi CBS 129742T MH865540 KM495395 − − Knoxdaviesia wingfieldii CBS 132470T NR_111723 NG_042656 − − Microascus longirostris AFTOL-ID 1237 − − DQ471026 − Nimbospora effusa JKI 5104A − U46892 U46877 DQ836887 Parascedosporium tectonae CBS 127.84T MH861707 EF151332 U43907 − Pseudoscopulariopsis schumacheri CBS 435.86T LM652455 AF400874 − − Synnematotriadelphia stilboidea CBS 101294 MF434783 MF434793 MF434811 MF434802 Synnematotriadelphia stilboidea CBS 221.85T MF434781 MF434792 MF434810 MF434801 Triadelphia disseminata CBS 138592T MF434784 MF434788 MF434806 MF434797 Triadelphia diversa CBS 113.90T MF434782 MF434790 MF434808 MF434799 Triadelphia fusiformis MFLUCC 16-0231T MH777097 MH777098 − − Triadelphia heterospora CBS 222.83T MF434779 MF434789 MF434807 MF434798 Triadelphia hexaformispora TBRC 9288T MK588842 MK588850 MK588840 MK578528 Triadelphia loudetiae CBS 589.77T MF434776 MF434785 MF434803 MF434794 Triadelphia moubasheri AUMC 10746T KY611849 − − − Triadelphia mukdahanensis MFLUCC 23-0049 T OQ871494 OQ871495 OQ883648 OQ871561 Triadelphia pulvinata CBS 590.77T MF434777 MF434786 MF434804 MF434795 Triadelphia pulvinata CBS 744.84 MF434780 MF434787 MF434805 MF434796 Triadelphia romanica CBS 162.79T MF434778 MF434791 MF434809 MF434800 Wardomyces anomalus CBS 299.61 LN850992 LN851044 − − 1 AFTOL-ID: Assembling the Fungal Tree of Life; AUMC: Assiut University Mycological Centre; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CMW: Collection of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, South Africa; JKI: Culture collection of the Julius Kühn-Institute, Siebeldingen, Germany; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; TBRC: Thailand Bioresource Research Center; T: ex-type isolates. 2 ITS: internal transcribed spacer regions 1 and 2 including 5.8S nrRNA gene; LSU: 28S large subunit of the nrRNA gene; SSU: 18S small subunit of the nrRNA gene; RPB2: partial RNA polymerase II second largest subunit gene. -
The combined dataset of ITS, LSU, SSU, and RPB2 sequence data including our new strain were analyzed by maximum likelihood (ML) and Bayesian analyses. The combined sequence alignment is comprised of 37 taxa (3,520 characters with gaps) with Falcocladium sphaeropedunculatum (CBS 111292) and F. multivesiculatum (CBS 120386) as outgroup taxa. A best scoring RAxML tree with a final likelihood value of −20,343.554289 is presented in Fig. 1. The matrix had 1,454 distinct alignment patterns, with 40.85% of undetermined characters. Estimated base frequencies were as follows: A = 0.249759, C = 0.237431, G = 0.277058, T = 0.235751; substitution rates: AC = 1.533582, AG = 3.298266, AT = 1.550122, CG = 1.111961, CT = 6.988888, GT = 1.000000; gamma distribution shape parameter α = 0.206975. In a BLASTn search of NCBI GenBank, the closest matches of the closest matches of the LSU, SSU and RPB2 sequence of Triadelphia mukdahanensis (MFLUCC 23-0049, ex-holotype) is T. heterospora (strain CBS 222.83) with 98.15% (MF434789), 99.85% (NG_067662), and 92.02% (MF434798) similarity, in respectively, while the closest matches of the ITS sequence with 91.86% similarity, was T. loudetiae (strain CBS 589.77, MF434776). Based on the phylogram generated from maximum likelihood analysis (Fig. 1) shows that our strain is closely related to T. heterospora with high bootstrap support (97% ML and 1.00 BYPP) and clustered with T. loudetiae. Interestingly, two Synnematotriadelphia stilboidea strains which introduced by Chuaseeharonnachai et al.[2] form a well-supported separate clade with Triadelphiaceae and Graphiaceae, with high bootstrap support (99% ML and 1.00 BYPP).
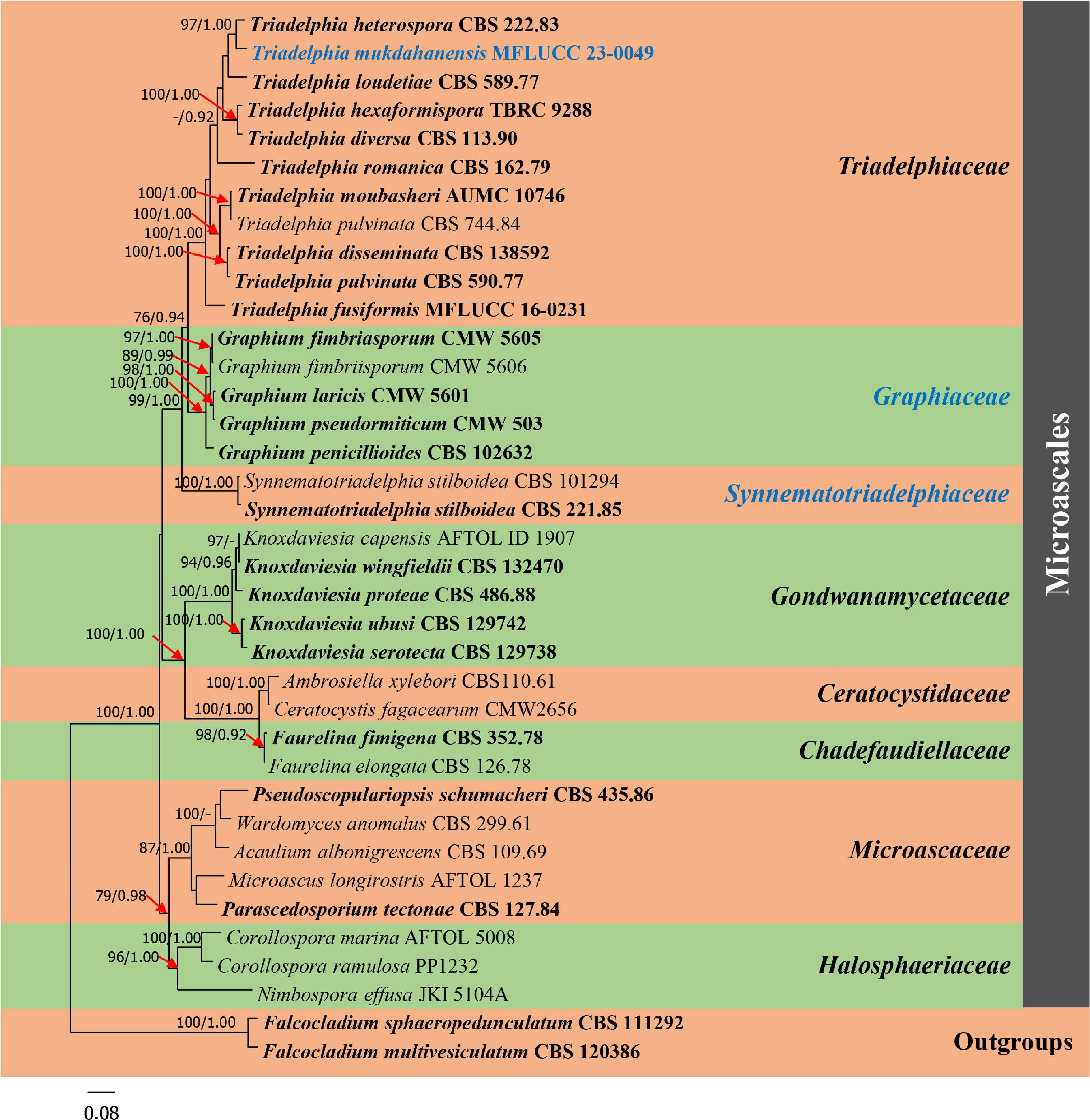
Figure 1.
Phylogram generated from maximum likelihood analysis based on combined dataset of ITS, LSU, SSU and RPB2 sequence data of Microascales group. Bootstrap support values for maximum likelihood (ML) equal to or greater than 75% and Bayesian posterior probabilities (PP) equal to or greater than 0.90 are given above the nodes. A newly generated sequence and a newly introduced family are in blue bold. Type species are in bold. The tree is rooted with Falcocladium sphaeropedunculatum (CBS 111292) and F. multivesiculatum (CBS 120386).
Taxonomy
-
Graphiaceae Z.W. de Beer, Seifert & M.J. Wingf. ex Mapook, Boonmee & K.D. Hyde, fam. nov.
Index Fungorum number: IF 559717, Faces of fungi number: FoF 01099
Type genus: Graphium Corda, Icon. fung. (Prague) 1: 18 (1837)
Saprobic on plant and wood substrates, occasionally isolated from soil, manure, polluted water, sometimes causing wounds on tree bark or associated with beetles, and fungemia in an immunosuppressed child post stem-cell transplant. Sexual morph: Undetermined. Asexual morph: Hyphomycetous. Colonies on the natural substrate superficial, Conidiomata synnematous, determinate with pigmented stipes. Conidiophores macronematous, penicillately branches in two to three levels, compact, pigmented, septate with metulae present at the apex. Conidiogenous cells in whorls of two to six, enteroblastic, phialidic, with annellidic extensions. Conidia cylindrical to obovoid, hyaline, aseptate, sometimes slightly curved with age, truncate at base, often with a distinct basal frill, conidial mass produced in a transparent and slimy droplet. Rarely synanamorphic forming obovoid, monoblastic, with pigmented chlamydospore-like conidia (adapted from de Beer et al.[6], Maharachchikumbura et al.[19], Hyde et al.[3]).
Type species: Graphium penicillioides Corda, Icon. fung. (Prague) 1: 18 (1837)
Holotype: CZECH REPUBLIC, Prague, on Populus nigra cv. ilalica, 14 December 1988, PR 155518; a slide from the holotype (DAOM 51800, isotype)
Epitype: CZECH REPUBLIC, České Budějovice, isolated from wood core of Populus nigra cv. ilalica, 3 September 1998, T. Kirisits, PRM 842988 (JCM 10498 = CBS J02632 = T. Kirisits No.3)
Morphological description and illustration: see Okada et al.[20] and Hyde et al.[3]
Notes: Graphiaceae was published by de Beer et al.[6] with nine accepted Graphium species based on morphological description of a type species, G. penicillioides and available DNA sequence data from other know species[20−22]. However, because no registration identifier was cited in the protologue, the family name is invalid (Nom. inval., Art. F.5.1)[7]. In this study, Graphiaceae form a distinct family in the clade comprising Triadelphiaceae and Synnematotriadelphiaceae within Microascales (Fig. 1). Therefore, we validate Graphiaceae as a separate family in the order Microascales to accommodate the genus Graphium by providing a registration identifier and presenting the corrected type citations along with references to the original descriptions herein.
Synnematotriadelphiaceae
Mapook & K.D. Hyde, fam. nov. Index Fungorum number: IF 559716, Faces of fungi number: FoF 14113
Type genus: Synnematotriadelphia Chuaseehar., Somrith., Nuankaew & Boonyuen, in Chuaseeharonnachai et al., Mycol. Progr. 19(2): 132 (2020)
Saprobic on a decayed petiole of a palm. Sexual morph: Undetermined. Asexual morph: Hyphomycetous. Colonies on natural substrata effuse, hairy, developing synnemata or hairy-like structures in synthetic media. Mycelium partly superficial, hyaline to subhyaline, or olivaceous, septate, branched hyphae. Conidiomata synnematous, erect, simple or branched, solitary to gregarious, determinate, cylindrical or clavate, brown to blackish brown; parallel hyphae of synnemata septate, pale brown, with terminal short branches bearing single or fasciculate conidiogenous cells. Conidiogenous cells arising from metulae on the side of synnemata, holoblastic, monoblastic, solitary or aggregated, oblong to clavate, or spherical to subspherical, subhyaline to olivaceous. Conidia pleomorphic, solitary, acrogenous, produced in two or three different conidial forms on synnemata [as conidial type (a), (c) and (e) in Triadelphia], septate with transverse septum or septa, hyaline to pigmented, thin- to thick-walled (adapted from Chuaseeharonnachai et al.[2]).
Type species: Synnematotriadelphia stilboidea (Mercado & R.F. Castañeda) Chuaseehar., Somrith., Nuankaew & Boonyuen, in Chuaseeharonnachai et al., Mycol. Progr. 19(2): 133 (2020)
Holotype: CUBA, Havana, Botanical Garden, on dead leaf petiole of Roystonea regia, 20 April 1982, A. Mercado, No. 6865, ex-holotype living culture CBS 221.85.
Morphological description and illustration: see Chuaseeharonnachai et al.[2]
Notes: Chuaseeharonnachai et al.[2] introduced a genus Synnematotriadelphia to accommodate two synnematous Triadelphia species (T. stilboidea and T. synnematofera). Two Synnematotriadelphia formed a basal taxa to Triadelphia and were positioned in Triadelphiaceae[2]. In our analyses, Synnematotriadelphiaceae forms a distinct family within Microascales in the clade comprising Triadelphiaceae and Graphiaceae (Fig. 1), with 99% ML and 1.00 BYPP bootstrap support. The three families differ as follows: Triadelphiaceae (type Triadelphia) has sporodochium-like structures, lack of conidiophores, conidia with one to five different forms, while Graphiaceae has synnematous, penicillately branched conidiophores, with cylindrical to obovoid, aseptate, hyaline conidia and Synnematotriadelphiaceae (type Synnematotriadelphia) has synnematous conidiophores with two or three different conidial forms on synnemata [as conidial type (a), (c) and (e) in Triadelphia].
Triadelphiaceae Y.Z. Lu, J.K. Liu, Z.L. Luo & K.D. Hyde, in Luo et al., Fungal Diversity: 10.1007/s13225-019-00438-1, [105] (2019)
Index Fungorum number: IF 555668, Facesoffungi number: FoF 05449
Morphological description and illustration: See Luo et al. (2019)
Type genus: Triadelphia Shearer & J.L. Crane, Mycologia 63(2): 247 (1971)
Triadelphia mukdahanensis Mapook, Huanraluek, Boonmee & K.D. Hyde, sp. nov. Fig. 2

Figure 2.
Triadelphia mukdahanensis (holotype). (a)–(d) Appearance of colonies on substrate. (e)–(g) Conidiogenous cells and conidia (e and f mounted in 5% KOH). (f)–(k) conidial type (a). (l)–(n) Conidial type (b). (o)–(p) Conidial type (f). (q) Conidial type (a), (b) and (f). Scale bars: a–c = 500 µm, d = 20 µm, e–i, k–n, q = 10 µm. j, o, p = 5 µm.
Index Fungorum number: IF 900159, Faces of fungi number: FoF 14114
Etymology: Referring to the location where the specimen was collected, Mukdahan Province, Thailand.
Holotype: MFLU 23-0071
Saprobic on decaying wood. Sexual morph: Undetermined. Asexual morph: Hyphomycetous. Colonies on the natural substrate superficial, sporodochium-like structures, scattered, thinly diffuse, black. Mycelium partly superficial and partly immersed in the substratum, smooth-walled, with septate hyaline hyphae. Conidiogenous cells holoblastic, monoblastic, integrated, borne directly on the mycelium, globose to ampulliform, hyaline to light brown. Conidia solitary, smooth-walled, pleomorphic, forming dark brown to pale olivaceous when mounted in 5% KOH, three different forms (a, b, & f); form (a): (9.5–)18–25 × (3.5–)5–7(–8.5) µm (
$\overline \chi $ $\overline \chi $ $\overline \chi $ Culture characteristics: Conidia germinating on MEA within 24 h at 25 ºC and germ tubes produced from the ends. Colonies on MEA irregular, mycelium slightly raised, undulate, greyish to olivaceous-brown at the surface, dark olivaceous in reverse.
Material examined: THAILAND, Mukdahan Province, Wat Pa Sanam Thong, Phang Daeng, Dong Luang District, on decaying wood, 24 July 2019, S. Boonmee, (MFLU 23-0071, holotype), ex-type culture MFLUCC 23-0049.
Notes: Triadelphia mukdahanensis (MFLUCC 23-0049) was collected from a terrestrial habitat. Based on phylogenetic analysis, T. mukdahanensis is closely related to T. heterospora with 97% ML and 1.00 BYPP bootstrap support. Triadelphia mukdahanensis have overlapping size of conidial forms type (a) and (f) with T. heterospora and both lack conidial forms type (c) (Table 2). However, T. mukdahanensis is different from T. heterospora in having three different conidial forms type (a), (b) and (f), lacking of conidial forms type (d) and (e), while T. heterospora has four different conidial forms type (a), (d), (e) and (f) but lacks conidial forms type (b) (Table 2). In addition, a comparison of the ITS (+ 5.8S) gene region of T. mukdahanensis with T. heterospora reveals 33 base pair differences (5.6%) across 591 nucleotides, while RPB2 gene reveals 71 base pair differences (8%) across 890 nucleotides (not include gaps).
Table 2. Conidial forms comparison of Triadelphia mukdahanensis and T. heterospora discussed in this study.
Conidial forms T. mukdahanensis
(this study)T. heterospora
(Shearer & Crane[23])T. heterospora
(Constantinescu & Samson[24])Type (a):
oblong to cylindrical, brown to dark brown, (1–)2–septate, septum covered with dark band(9.5–)18–25 ×
(3.5–)5–7(–8.5) µm(14.7–)16.2–19(–21.2) ×
3.5–6 µm15–23 × 5–8.5 µm Type (b):
pyriform to broadly clavate, or club-shaped, brown to dark brown, (1–)2–septate, the apical septum covered with broad dark bands, the basal cell subhyaline or light brown to brown20–30 × 7–12 µm – – Type (c):
obclavate to acicular with a narrow long tip, hyaline or yellowish brown, multiseptate– – – Type (d):
fusiform, obclavate with acicular tips, or rounded at the tip, multi-septate, end cells pale brown, central cells brown to dark brown, dark band covering over the central septa– 17.6–23.5 × 8.2–10.6 µm 17.6–23.5 × 8.2–10.6 µm Type (e):
allantoid or reniform, hyaline or pale yellowish, 0–3-septate– – 8.5–15 × 3–5 µm Type (f):
obovate to broadly ellipsoidal, pale brown, unicellular5–9 × 3.5–5 µm – 6–8 × 4–5 µm -
Triadelphia is mostly found in terrestrial and rarely in freshwater habitats[2,8]. Based on our phylogenetic results, our new strain grouped within Triadelphia and is closely related to T. heterospora. Simultaneously, we compared our new strain with the 20 epithets listed in Index Fungorum[7], and found that our new strain has the most morphological similarity to T. heterospora. Triadelphia heterospora was originally reported with two different conidial forms (a) and (d)[23]. Subsequently, Constantinescu & Samson[24] reported two additional conidial forms (e) and (f). Although our new strain has overlapping size of conidial forms type (a) and (f) with T. heterospora and both are lacking conidial forms type (c), our new strain is different in having three conidial forms type (a), (b) and (f), lacking conidial forms type (d) and (e) while T. heterospora have lacking reported of conidial forms type (b), which is appeared in our new strain. Therefore, we introduced a new species, T. mukdahanensis based on both morphology and phylogeny.
Interestingly, Synnematotriadelphia strains, which were positioned in Triadelphiaceae in Chuaseeharonnachai et al.[2], form a well-supported separate clade to Triadelphiaceae and Graphiaceae within Microascales, based on combined dataset of ITS, LSU, SSU and RPB2 sequence data. Therefore, we introduce a new family, Synnematotriadelphiaceae to accommodate two Synnematotriadelphia species (S. stilboidea and S. synnematofera), based on both morphology and phylogeny. Synnematotriadelphiaceae (type Synnematotriadelphia) has synnematous conidiophores with two or three different conidial forms on the synnemata as conidial type (a), (c) and (e) in Triadelphia[2], while Triadelphiaceae (generic type Triadelphia) has sporodochium-like structures, lack of conidiophores with one to five different types of conidial forms[8−10], and Graphiaceae has synnematous with penicillately branched conidiophores, with cylindrical to obovoid, aseptate, hyaline conidia[3, 6, 19]. This can be indicated that Synnematotriadelphiaceae have shared some characteristics from both Triadelphiaceae and Graphiaceae. However, Graphiaceae, which was introduced by de Beer et al.[6], is recorded as an invalid name in Index Fungorum[7] due to an identifier issued by a recognized repository was not cited in the protologue. In compliance with Article F.5.1[25], Graphiaceae, is registered in Index Fungorum[7] and obtained with a unique number. The family is validated by providing a registration identifier and presenting the corrected type citations along with references to the original descriptions herein.
Ausana Mapook would like to thank the Mae Fah Luang University Fund (Grant No. 651A16029), entitled 'Taxonomy, phylogeny, risk assessment, and potential impact of fungi on Siam weed in northern Thailand'. Kevin D. Hyde thanks the Basic Research Fund supported from National Science, Research and Innovation Fund (Grant No. 652A01001), entitled 'Studies of fungi associated with Asteraceae and the discovery of biological properties', Princess Srinagarindra's Centenary Celebrations Foundation (Grant No. 64316001), and National Research Council of Thailand (NRCT) grant, entitled 'Total fungal diversity in a given forest area with implications towards species numbers, chemical diversity and biotechnology' (Grant No. N42A650547). He also thanks Chiang Mai University for the award of Visiting Professor. Saranyaphat Boonmee would like to thank the Mae Fah Luang University Fund (Grant No. 631C15001) and Plant Genetic Conservation Project under the Royal Initiation of Her Royal Highness Princess Maha Chakri Sirindhorn-Mae Fah Luang University.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Mapook A, Hyde KD, Huanraluek N, Boonmee S. 2023. Addition to Microascales (Sordariomycetes, Ascomycota): Synnematotriadelphiaceae fam. nov., Triadelphia mukdahanensis sp. nov. (Triadelphiaceae) and the validation of Graphiaceae. Studies in Fungi 8:10 doi: 10.48130/SIF-2023-0010
Addition to Microascales (Sordariomycetes, Ascomycota): Synnematotriadelphiaceae fam. nov., Triadelphia mukdahanensis sp. nov.
- Received: 02 February 2023
- Accepted: 03 April 2023
- Published online: 15 May 2023
Abstract: A new species, Triadelphia mukdahanensis from northeastern Thailand, is introduced based on morphology and multi-gene phylogeny of combined ITS, LSU, SSU, and RPB2 sequence datasets. Phylogenetic analysis indicates that T. mukdahanensis is closely related to T. heterospora with high bootstrap support. However, T. mukdahanensis is distinct from T. heterospora and other exist species in conidial characteristics. In addition, the present study clarifies the current taxonomic placement of the genus Synnematotriadelphia based on phylogenetic analyses. The result shows that Synnematotriadelphia forms a distinct lineage in the clade comprising Triadelphiaceae and Graphiaceae. Therefore, a new family, Synnematotriadelphiaceae, is introduced to accommodate this distinct lineage and Graphiaceae is also validated herein.
-
Key words:
- 1 new species /
- Graphiaceae /
- Northeastern Thailand /
- Phylogeny /
- Synnematotriadelphiaceae /
- Taxonomy












