-
Papaya (Carica papaya L.) is a tall herbaceous fruit plant from the Caricacae family[1]. It is believed to be native to tropical regions of the Americas and widely cultivated in all tropical regions of the world due to its high nutritional and economic value[2]. Papaya fruits are a rich source of vitamins A and C, folate, thiamine, niacin, riboflavin, iron, potassium, calcium, and fiber. The entire papaya plant, including seeds, leaves, roots, and flowers, has long been used for medicinal purposes owing to its anti-inflammation, anti-skin aging, wound healing, gastrointestinal health improving, and cancer risk-reducing properties[3,4]. In addition, the protease papain, the primary ingredient of which is papaya latex from raw fruits, is widely used in food processing, medicine, and cosmetics[3,5].
In the past decade, the market demand for papaya has increased steadily due to rapid population growth and the improved public perception of the health benefits of functional fruits. According to the latest statistics from the Food and Agriculture Organization, the world production of papaya fruits has increased from 10.8 million metric tons (Mt) in 2010 to 13.6 Mt in 2020. The global harvested area has grown from 400,200 hectares in 2010 to 468,700 hectares in 2020. India is the world's largest producer of papaya in 2020, with 6.01 Mt of fruit, accounting for 43.2% of the total global production. Other major papaya-producing countries include Dominican Republic, Brazil, Mexico, Indonesia, Nigeria, Democratic Republic of Congo, Colombia and Thailand. Mexico and the United States are the leading papaya-exporting and -importing countries, respectively. However, some biotic stresses (pathogens, pests, and weeds), abiotic stresses (extreme temperatures and drought), and post-harvest losses caused by pathological, physiological, and mechanical conditions have been severely impacting global production of papaya[1]. Traditional papaya breeding methods are limited due to the lack of natural genetic resources and a small gene pool since papaya is the only species of the genus Carica[6]. Transgene technologies provide a powerful alternative to conventional methods for generation of modified plants with the desired traits. Transgenic papaya with resistance to papaya ringspot virus (PRSV, genus Potyvirus, family Potyviridae) was the first genetically modified fruit crop for eventual commercial production in 1998 in Hawaii[7]. Since then, papaya has become the main tropical crop for biotic and abiotic stress tolerance and fruit quality improvement via genetic engineering[8,9]. After the completion of the papaya genome sequence in 2008[10], the recently established genome editing based on the CRISPR-Cas systems will open a new era of precision breeding of papaya[11,12]. This review provides an overview of past and recent gene transformation methods and their applications in papaya breeding (Fig. 1, Table 1). Additionally, the paper discusses the future perspectives and challenges of papaya molecular breeding (Fig. 1).
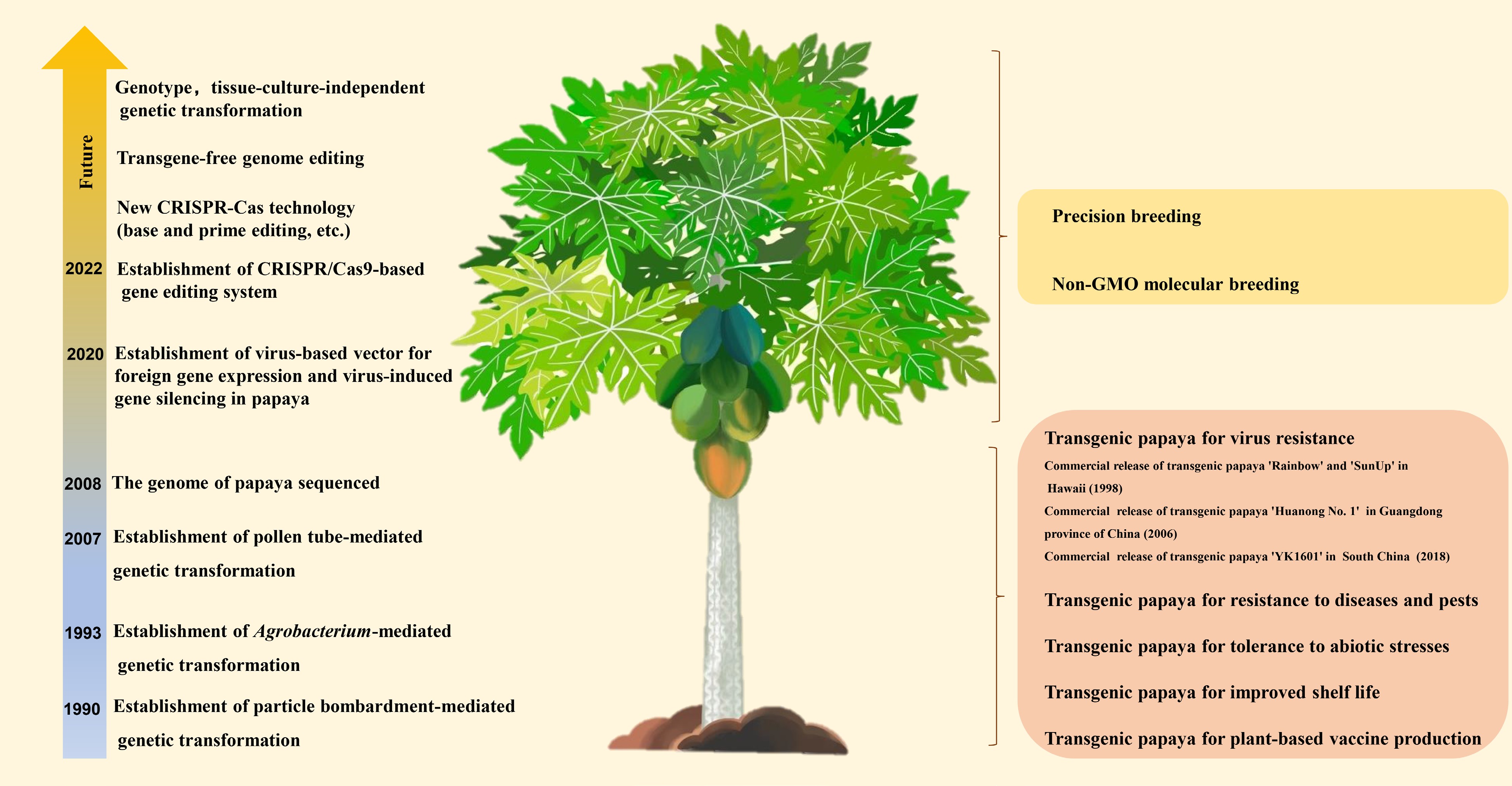
Figure 1.
Important historical milestones and future perspectives in gene transformation strategies and their application of Carica papaya L.
Table 1. Summary of transgenic studies in papaya genetic improvement.
Trait improvement Gene introduced Transformation method Reference Virus resistance PRSV resistance Translatable/ Untranslatable coat protein (CP) Biolistics [15,16,39−42,44,45] Translatable/ Untranslatable CP Agrobacterium [21,24,25,34,39,
41−43, 46−48]Replicase Agrobacterium [34] Untranslatable CP Pollen tube [30] PRSV and PLDMV resistance Untranslatable CP Agrobacterium [26,52] Fungal resistance Phytophthora palmivora resistance Stilbene synthase Vst1 Biolistics [53] Antimicrobial peptide DmAMP1 Biolistics [54] NPR1 Biolistics [55] Bacterial resistance Dieback disease resistance Acyl-homoserine lactonase Agrobacterium [61] Insect resistance Carmine spider mite resistance Chitinase Biolistics [62] Lectin GNA Biolistics [66] Abiotic stresses tolerance Herbicide resistance Bar Biolistics [17] Aluminum tolerance Citrate synthase Biolistics [68] Improvement of shelf life Untranslatable ACC oxidase (ACO) Biolistics [72] Untranslatable ACO1 and ACO2 Agrobacterium [60,73] Untranslatable ACC synthase (ACS) Biolistics [9,74] Plant-based vaccine Antituberculosis vaccine ESAT6 of tuberculosis Agrobacterium [81] Anticysticercosis vaccine Synthetic peptide genes (KETc1, KETc12, KETc7) Biolistics [82,84] -
The particle bombardment method, also known as biolistics, is a technique that uses high-velocity microprojectiles made of gold or tungsten to deliver foreign DNA to regenerable plant cells and tissues via a gene gun[13]. In 1990, Fitch et al. reported the first successful stable transformation of papaya using microprojectile bombardment[14]. They used tungsten particles to bombard immature zygotic embryos, hypocotyl sections, and somatic embryos from Hawaiian cultivars 'Sunset' and 'Kapoho' with a plasmid carrying a selectable marker (neomycin phosphotransferase II, nptII) and a reporter gene (β-Glucuronidase, gus). The resulting transgenic embryos and papaya shoots stably expressed the chimeric genes coding for NPTII and GUS. Fitch et al. also obtained PRSV-resistant R0 transgenic plant lines by using particle bombardment-mediated transformation of 2,4-D-treated 'Sunset' immature zygotic embryos with a vector carrying the translatable coat protein gene (CP) from PRSV Hawaiian mild strain HA 5-1[15]. To improve transformation and plant regeneration efficiencies after bombarding papaya somatic embryos, Gonsalves et al. developed three modified major procedures including the culture of suitable somatic embryos at the time of bombardment, efficient proliferation of transformed embryos without antibiotics at the later stages, and selection of plantlets 0.5–1.0 cm long at the transplant stage[16]. On average, each somatic embryo cluster generated 1.3 transgenic lines expressing the untranslatable CP gene of PRSV HA 5-1 in 'Sunrise'. In 1995, Cabrera-Ponce et al. developed herbicide-resistant transgenic papaya plants using zygotic embryos and embryogenic callus of Mexican variety 'Maradol' via particle bombardment[17]. Bombarded embryogenic callus about 50 mg regenerated at least two transgenic clones, and the transformation efficiency was 50-fold higher than that reported by Fitch et al. The age and growth characteristics of the embryogenic callus was considered the most important factor affecting the biolistic transformation frequency. Furthermore, Zhu et al. found that papaya transformation efficiency by particle bombardment was significantly increased using visual green fluorescent protein (GFP) or phosphomannose isomerase (PMI)/mannose as the selection marker and system, instead of antibiotics[18]. Particle bombardment-mediated genetic transformation has been established in papaya cultivars from different countries, including Australia, Jamaica and Brazil, and applied for development of transgenic papaya with new traits such as disease resistance, fruit quality improvement and plant-based vaccine production (Table 1)[9,19]. However, this method has some disadvantages, such as expensive biolistic equipment and consumables, frequent multiple copies of integrated transgenes, transgene rearrangement, and the co-suppression phenomenon[20,21].
Agrobacterium-mediated genetic transformation
-
Compared with particle bombardment, Agrobacterium-mediated genetic transformation is widely-used method to deliver genes of interest into plants due to its lower transgene copy number, lower cost and higher efficiencies[22]. In 1988, Pang & Sanford were the first to transform leaf disks, stems and petioles of papaya via Agrobacterium, but failed to regenerate transformed plantlets[23]. Fitch et al. reported the successful production of transgenics papaya from Agrobacterium-mediated transformation of somatic embryos from hypocotyl sections[21]. However, the transformation frequency was low (0.15%) and only two transgenic lines were regenerated from 13 g of embryogenic callus. When papaya petioles of in vitro-propagated multishoots were used for Agrobacterium-mediated transformation, the transformation frequency was increased by about 3%[24]. In both cases, the total period of regeneration was long (10–13 months), limiting their application in producing transgenic papaya. To overcome this hurdle, Cheng et al. reported that wounding cultured embryogenic tissues with carborundum prior to Agrobacterium-mediated transformation achieved up to 15.9% transformation frequency and the transformation period is shortened to about 9 months[25]. Commercial markets prefer the pear-shaped fruit obtained from hermaphroditic plants. Accordingly, somatic embryos derived from adventitious roots of hermaphroditic plants were used as explants to generate hermaphrodite transgenic papaya lines with double resistance to PRSV and papaya leaf distortion mosaic virus (PLDMV genus Potyvirus, family Potyviridae) via Agrobacterium-mediated transformation[26]. This protocol contributes to develop new papaya varieties with desired hermaphroditic characteristic in a shorter period of time. In a further modification, Jiang et al. established a sonication-assisted Agrobacterium-mediated transformation in papaya[27]; the transformation frequency was enhanced about 10 times using 15 s sonication of embryo callus.
Pollen tube-mediated genetic transformation
-
Pollen tube-mediated genetic transformation is a non-tissue culture method that provides an alternative to overcome genotype specificity limitations[28]. Papaya is an ideal model for transformation via the pollen tube pathway because of its trioecious characteristic with male, female, and hermaphrodite flowers, short generation time (9–15 months), and continuous flowering and fruit production throughout the year with numerous seeds[29]. Wei et al. pipetted various binary plasmids or Agrobacterium suspensions carrying PRSV CP fragments in sense, antisense, and double-sense orientations onto stigmas of female flowers before pollination[30]. PCR analysis revealed that approximately 9% of the papaya seeds from treated flowers were transformed. The resulting transgenic plants expressed PRSV CP gene and exhibited the resistance to PRSV. This method provided a promising genetic transformation for directly generating transgenic papaya without the need for tissue culture.
-
PRSV is a devastating pathogen that causes significant yield losses in papaya plantations worldwide[31]. There are two types of PRSV, P and W; P infects papaya and cucurbits, while W infects cucurbits but not papaya[31]. Because of a lack of sources of natural genetic resistance to PRSV, the main control strategies are quarantine, physical control, cross-protection, and transgene-based resistance[7]. Currently, transgenic resistance conferred by homology-dependent post-transcriptional gene silencing (PTGS) of the viral gene is the most effective method to protect plants against PRSV infection. As early as 1991, an R0 transgenic line 55-1 resistant to PRSV HA was first obtained in the USA by particle bombardment transformation of the red-fleshed cultivar 'Sunset' with the CP gene of PRSV, based on the concept of pathogen-derived resistance[15]. Subsequently, homozygous 'SunUp' was produced by crossing line 55-1 carrying a single copy of PRSV CP and a non-transgenic 'Sunset', followed by selfing of the transgenic R1 progeny. Given that the yellow-flesh dominant cultivar 'Kapoho' was popular in Hawaii, line 55-1 was further used to cross with nontransgenic 'Kapoho' and the resulting F1 hybrid was named as 'Rainbow'[32]. In 1998, 'SunUp' and 'Rainbow' were released for commercial cultivation in Hawaii, and they helped revive the Hawaiian papaya industry[33]. This case represents the first commercialized transgenic fruit crop. In 2006, transgenic papaya 'Huanong No. 1' from the South China Agricultural University was approved for release in Guangdong province (China). This genetically modified papaya was generated using Agrobacterium-mediated transformation of the cultivar 'Yuanyou No.1' with the replicase (nuclear inclusion protein b) gene (NIb) of PRSV Ys, a prevalent strain in China[34]. In 2018, The Chinese Ministry of Agriculture and Rural Affairs issued biosafety certificates for commercial production of the transgenic papaya line YK1601 expressing the CP gene of the severe PRSV strain YK in South China. However, transgenic papaya is susceptible to reduction or loss of viral gene-mediated resistance to PRSV due to various factors such as transgene copy number, plant developmental stage, the sequence homology with the transgene, and the geographic diversity of PRSV[35−38]. Therefore, geographic-specific varieties of PRSV-resistant transgenic papaya have been developed using transgenes that expressed translatable or untranslatable sequences (sense, antisense RNA or hairpin RNA) of CP or NIb from different PRSV strains of various countries (Table 1)[9,19,34,39−48]. In addition, another potyvirus, PLDMV, was detected in a PRSV-resistant transgenic papaya in China, causing disease symptoms similar to those of PRSV[49,50]. Moreover, co-infection with PRSV and PLDMV was identified in papaya[51], making PLDMV an emerging threat to papaya production in China. Kung et al. transformed an untranslatable chimeric sequence including the truncated CP coding region of the PLDMV P-TW-WF isolate and the truncated CP coding region with the complete 3′ untranslated region of PRSV YK isolate into papaya, resulting in transgenic plants with double resistance to PLDMV and PRSV[52]. Moreover, to decrease the cycle time and costs in hybrid breeding, hermaphrodite transgenic papaya lines with resistance to both PRSV and PLDMV were generated via transforming somatic embryos derived from adventitious roots of in vitro shoots[26].
Transgenic papaya for resistance to diseases and pests
-
Phytophthora palmivora is an oomycete plant pathogen that causes rots in the roots, stems and fruits of papaya, and is highly destructive. Zhu et al. transformed a stilbene synthase (Vst1) from grapevine (Vitis vinifera L.), antimicrobial peptide 1 (DmAMP1) from seeds of dahlia (Dahlia merckii), or NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1) from Arabidopsis thaliana or papaya into papaya plants to confer resistance to P. palmivora[53−55]. Stilbene synthases (ST) are the key enzymes in the biosynthesis of the stilbene backbone, and transgenic plants overexpressing ST genes could improve resistance to pathogens that are sensitive to stilbenes[56] . Indeed, expression of the Vst1 gene in papaya increased resistance to P. palmivora[53]. DmAMP1 is an antifungal plant defensin that induces an array of relatively rapid responses in fungal membranes[57]. Overexpression of DmAMP1 in transgenic papaya lines increased their resistance to P. palmivora by reducing hyphal growth at the infection sites[54]. NPR1 is a well-known key regulator of systemic acquired resistance that can trigger plant defense after local infection by pathogens[58]. Transgenic papaya expressing NPR1 from Arabidopsis thaliana or papaya showed improved resistance to P. palmivora[55]. In Malaysia, dieback disease caused by Erwinia mallotivora is a major threat to papaya plantations[59]. Sekeli et al. transformed two acyl-homoserine lactonase genes with anti-pathogenic function from Bacillus cereus CHB37 and Bacillus thuringiensis SP24 into papaya. The resulting transgenic papaya lines exhibited resistance against the dieback disease[60, 61].
Carmine spider mite (Teiranychus cinnabarinus Boisduval) is one of the most destructive pests affecting papaya[62]. Chitinases play an important role in degrading chitin in the exoskeleton and gut linings of insects[63]. Several studies have demonstrated that transgenic plants overexpressing insect chitinases are protected from pathogenic fungi and insect pests, as chitinases in high concentrations or with appropriately timed exposure are toxic to plant pathogens such as insects and fungi[64,65]. McCafferty et al. developed insect-resistant transgenic papaya plants expressing a tobacco hornworm (Manduca sexta) chitinase (MSCH)[62]. Insect bioassays revealed that expression of the insect MSCH gene improved papaya tolerance to carmine spider mite. Furthermore, transgenic papaya plants overexpressing a snowdrop (Galanthus nivalis) lectin gene GNA were identified to contain biologically active lectin with spider mite control activity[66].
Transgenic papaya for tolerance to abiotic stresses
-
The bialaphos resistance (Bar) gene, which confers resistance to the broad-spectrum herbicide glufosinate via N-acetylation, has been widely used in genetically-engineered herbicide-resistant plants in many crop species[67]. Cabrera-Ponce et al. successfully introduced the Bar gene by particle bombardment into papaya zygotic embryos and embryogenic callus to produce herbicide-resistant transgenic plants[17]. The study provides not only a new selection marker for papaya genetic transformation but also offers the possibility of applying transgenic technology to control weeds in papaya fields. Another example of transgenic papaya for tolerance to abiotic stresses is the tolerance to aluminum (Al) stress. de la Fuente et al. reported that overexpression of a citrate synthase gene from Pseudomonas aeruginosa in papaya led to a 10-fold increase in citrate production and improved tolerance to Al-stress up to 300 mM[68,69].
Transgenic papaya for improved shelf life
-
Papaya is a typical climacteric fleshy fruit that undergoes fast pulp softening due to ethylene which results in postharvest losses and shortened shelf life[70]. Ethylene is synthesized from methionine through S-adenosyl-L-methionine (SAM) and 1-aminocyclopropane-1-carboxylic acid (ACC). ACC synthase (ACS) and ACC oxidase (ACO) are the key ethylene biosynthetic enzymes[71]. A transgenic gene-silencing approach to prevent fruit ripening is to down-regulate the expression of key ethylene biosynthetic enzymes for suppression of ethylene production or perception. To engineer papaya fruit with delayed ripening, transgenic papaya was first developed by introducing a fragment of the ACO gene in the sense orientation to inhibit ethylene production and delay ripening[72]. Similarly, Malaysian researchers applied hairpin RNA (hpRNA) constructs to down-regulate the expression of the endogenous ACO1 and ACO2 genes in Eksotika papaya[60,73]. Fruits harvested from transgenic papaya plants showed an extended post-harvest shelf life of 14−20 d compared with the non-transgenic Eksotika papaya. In the Philippines, transgenic papaya delayed fruit ripening was generated by introduction of antisense ACS gene. The safety assessment revealed that downregulation of the expression of ACS gene did not produce any major undesired changes in the proximate composition and other chemical constituents in transgenic papaya[9,74].
Transgenic papaya for plant-based vaccine production
-
Since the first production of human growth hormone in transgenic tobacco in 1986, plants have been used as bioreactors to produce a wide range of recombinant human or animal proteins, such as enzymes, antibodies, vaccines, and medicinal proteins by molecular farming[75,76]. Edible vaccines have been shown to induce both mucosal and systemic immunity against disease-causing organism[77]. Given the attractive advantages of plant-based expression systems, such as cost-effectiveness, low cost, and easy cultivation, many edible plant species, including papaya, have been developed to generate plant-based vaccines against various human and animal diseases[78−80]. In a study aimed at developing a plant-based vaccine against Mycobacterium tuberculosis in papaya, transgenic papaya lines expressing the 6 kDa early secreted antigen target (ESAT6) were generated[81]. The work presents a new method for the development of edible vaccines against tuberculosis. Additionally, papaya callus cells were engineered to express three protective peptides (S3Pvac: KETc1, KETc12, and KETc7) for producing an oral vaccine against cysticercosis caused by the tape worms Taenia solium or T. crassiceps via particle bombardment[82]. Oral vaccination of mice and pigs with the S3Pvac-papaya soluble extract elicited effective immunity against cysticercosis, demonstrating that papaya callus is a useful platform for plant-based oral vaccines[83]. Recently, cell suspension cultures of KETc7-expressing calluses have been developed for biomass production of antiparasitic product against the gastrointestinal nematode Haemonchus contortus in airlift bioreactor[84].
-
Plant virus-based vectors are rapid and effective genetic tools without the need for time-consuming transformations and have been used for heterologous gene expression, endogenous gene silencing, and genome editing in plant functional genomic studies and biotechnology[85−87]. We successfully constructed two agroinfection-compatible fluorescence-tagged PLDMV infectious cDNA clones, driven by the Cauliflower mosaic virus 35S promoter, using a one-step Gibson assembly to express systematically GFP or mCherry protein in papaya plants[88,89]. The availability of these infectious clones will facilitate tracking of the dynamics of virus infection in plants and contribute to studies on PLDMV-host interactions. In the future, PLDMV-mediated overexpression vector will be used to produce high levels of recombinant proteins and peptides for applications in biopharmaceuticals and agrobiotechnology. Although the genome of papaya has been sequenced[10], the genetic approaches available at present for functional genomic studies depend on laborious genetic transformation. Virus-induced gene silencing (VIGS) is a powerful forward and reverse genetic technique for functional genomic studies in plants. More than 50 VIGS vectors from plant viruses and their viral satellites have been developed so far[90]. Recently, we developed a VIGS vector in papaya using a PLDMV mild strain, namely PLDMV-E[91]. The PLDMV-based VIGS vector successfully silenced five papaya endogenous genes, which encode phytoene desaturase (PDS), Mg-chelatase H subunit, putative GA receptors GIBBERELLIN (GA)-INSENSITIVE DWARF1A and 1B, and the cytochrome CYP83B1, respectively. The method provides a rapid and convenient tool as an alternative to transgenic techniques for functional genomic studies in papaya.
-
Over the past decade, genome editing techniques based on sequence-specific engineered endonucleases (SSNs), such as zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/associated (Cas) proteins, have been used as powerful tools for targeted genome modifications in plants[80,92−95]. SSNs generate double-stranded DNA breaks that are repaired in eukaryotic cells by two main types of DNA repair machinery, non-homologous end joining and homology-directed repair[93,96−98]. Among these tools, the CRISPR/Cas9 system is the most-widely applied genome-editing tool for genetic studies, germplasm innovation and crop breeding because of its ease, convenience, cost-effectiveness and high efficiency in creating targeted modifications. Given the availability of papaya genetic transformation technologies and whole-genome sequences, it is now feasible to develop CRISPR-based genome editing for papaya breeding. Recently, Brewer & Chambers established an effective CRISPR/Cas9-based gene editing system in papaya by expressing three guide RNAs (gRNAs) targeting the papaya PDS gene using Agrobacterium-mediated transformation[12]. Edited transgenic plantlets exhibited complete albino phenotypes, and diverse gene editing, including insertion, inversion, and deletion, was detected at all three desired gRNA target loci with an 81% efficiency of insertion or deletion (indel) mutations. Additionally, Hoang et al. developed an in vivo hairy root system for assessing genome editing efficiency in papaya via Rhizobium rhizogenes[11]. These studies have opened a new avenue for functional analysis of papaya genes, trait improvement, and breeding using the CRISPR/Cas9 system.
-
For the past 20 years, transgenic technology has proven successful in improving various agronomic traits in papaya, such as fruit quality, and biotic and abiotic stress tolerance (Fig. 1, Table 1). Notably, genetically engineered papaya expressing the PRSV CP is the first fruit crop to be successfully deregulated and commercialized. However, the development of genetic engineering in papaya lags behind that in many staple food crops and fruits over recent years. The main reasons are that papaya transformation is still dependent on traditional Agrobacterium-mediated genetic transformation or particle bombardment, both with some disadvantages such as low transformation efficiencies and laborious procedures. To improve papaya transformation systems, previous studies focused on optimizing the crucial transformation factors and plant regeneration conditions, such as explant types, tissue culture media and selection markers. In addition, current transformation protocols are only applicable to a limited number of papaya cultivars. In the future, more efforts are needed to develop simpler and more effective technologies and strategies for the improvement of papaya transformation and regeneration. Several studies have provided evidence that co-delivering transgenes with genes encoding some developmental regulators, such as WUSCHEL (WUS), Baby Boom (Bbm) and GROWTH-REGULATING FACTOR (GRF), can promote somatic embryogenesis and transformation efficiency in monocot genetic transformation[99−102]. For dicot species, Dr. Voytas' lab recently developed two transformation methods, named direct delivery (DD) and fast-treated Agrobacterium co-culture (Fast-TrACC), by co-delivering developmental regulators (WUS2, IPT and STM) and gene editing reagents into somatic cells to induce de novo genetically modified meristems in seedlings grown in nonsterile conditions and soil[103,104]. Transgenic shoots with targeted DNA mutations were recovered from the edited meristems of tomato, potato and grape. This innovative work avoids tissue culture-based transformation and provides simple and rapid alternatives to genetic transformation methods. More recently, Cao et al. established a simple Agrobacterium rhizogenes-mediated cut-dip-budding (CDB) delivery system to create transformed roots that produce transformed buds by root suckering[105]. This system has been used successfully to generate heritable transformation in two herbaceous plants (Taraxacum kok-saghyz and Coronilla varia), a tuberous root plant (sweet potato), and three woody plant species (Ailanthus altissima, Aralia elata, and Clerodendrum chinense). In addition, nanotechnology-based gene-delivery methods have been developed for plant genetic transformation and show many advantages over conventional methods[106]. In any case, these breakthrough technologies can be expected to contribute to developing new gene transformation strategies for crops including papaya, with applications in both biological research and genetic improvement.
The rapid advancement of CRISPR/Cas-based genome editing methods has revolutionized crop improvement and provided a powerful tool for enhancing various traits in papaya, such as resistance to pathogens and pests, abiotic stress tolerance, and fruit quality[95,107,108]. Although the CRISPR/Cas9-mediated genome editing system has been established in papaya, its application is still in the early stages compared to model plants and staple food crops. With the continuous innovation in CRISPR/Cas9 -based genome editing technology, novel techniques, more precise and efficient techniques, such as alternative types of Cas effectors-based gene editing, base editing, and prim editing, are emerging[94,109,110]. The novel techniques will further facilitate the development of precision breeding of papaya in the future (Fig. 1). In addition, the use of CRISPR/Cas-mediated gene editing raises important legislative issues related to genetically modified organisms (GMO), such as off-target mutations and transgene integration from T-DNA-mediated CRISPR/Cas9 gene editing. Transgene-free genome editing could address these concerns and enable regulatory approval for commercial production because the resulting mutation events without foreign DNA insertion could occur naturally or by conventional breeding. Some countries, such as the USA, Japan, and Australia, exclude some or all types of genome-edited crops from GMO regulations if they do not contain genetically engineered transgenes or foreign DNA[111,112]. Currently, the main strategies for eliminating CRISPR constructs and other transgenes fall into three categories[113−115]: (1) efficient elimination of transgenes from stable integration by genetic segregation, (2) transient expression of CRISPR/Cas DNA vectors and (3) direct delivery of CRISPR/Cas RNA or ribonucleoproteins. We anticipate that these new strategies of transgene-free genome editing will contribute to future development of edited papayas for commercial exploitation.
This work was supported by the National Natural Science Foundation of China (32072390), the Hainan Provincial Natural Science Foundation of China (320RC717), the Central Public Interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (grant no. 19CXTD-33) and the Innovation Platform for Academicians of Hainan Province.
-
Wentao Shen is the Editorial Board member of Journal Tropical Plants. He was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of this Editorial Board member and his research groups.
-
Received 11 January 2023; Accepted 17 April 2023; Published online 26 April 2023
-
# These authors contributed equally: Decai Tuo, Chunhua Ma
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press on behalf of Hainan University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Tuo D, Ma C, Yan P, Kong H, Zhou P, et al. 2023. Genetic transformation and gene delivery strategies in Carica papaya L.. Tropical Plants 2:5 doi: 10.48130/TP-2023-0005
Genetic transformation and gene delivery strategies in Carica papaya L.
- Received: 11 January 2023
- Accepted: 17 April 2023
- Published online: 26 April 2023
Abstract: Papaya (Carica papaya L.) is a popular tropical fruit of high commercial value due to its nutritional, medicinal and industrial properties. However, conventional breeding methods for papaya have several limitations, including a scarcity of natural genetic resources in the Carica genus and sexual incompatibility among closely-related species. To overcome these challenges, particle bombardment, Agrobacterium-mediated transformation, and pollen tube-mediated transformation have been developed for generating genetically modified papaya with the desired traits over the past three decades. These transformation methods have been successfully employed to improve papaya's resistance to biotic and abiotic stress, fruit quality, and even to produce edible vaccines. Notably, papaya ringspot virus-resistant transgenic papaya is the first successful commercial application of genetic engineering technology in a fruit crop. Recently, CRISPR/Cas9-mediated genome editing technology and viral vector-based gene delivery system have emerged as promising tools for papaya genetic improvement and functional genomic studies in papaya. Genome editing will open a new era of precision breeding for papaya. This review aims to highlight past and recent gene transformation methods and their applications in papaya breeding, as well as future perspectives and challenges in improving papaya via genetic engineering.
-
Key words:
- Gene transformation /
- Papaya /
- Transgene /
- Genetic engineering /
- Genome editing












