-
Umami has played an essential role in the Chinese diet since ancient times. References to umami can be found in many classical Chinese texts dating back to early history. For instance, during the Song Dynasty (960 AD to 1279 AD), Hong Lin mentioned in 'Shanjia Qing Offering' that bamboo shoots 'taste very fresh', and Yizun Zhu of the Ming Dynasty (1368 AD to 1644 AD) also noted in 'The Secret of Food Constitution' that 'pickled pork had a special umami taste'. These references reflect the historical appreciation and pursuit of umami by the Chinese people, although they were based on subjective perceptions at that time. It was not until Professor Ikeda's groundbreaking work that umami was studied as a scientific issue for the first time. He isolated glutamate from kelp (Laminaria japonica) and discovered its ability to enhance soup flavor. This unique taste, distinct from sour, sweet, bitter, and salty flavors, led Professor Ikeda to coin the term 'umami'[1,2]. However, due to limited systematic scientific research on umami at that time, it was not recognized as a basic taste by the scientific community; instead, it was considered a comprehensive taste associated with pleasure and increased appetite. It wasn't until 2002 when umami receptors (mGlu1/4, T1R1/T1R3) were discovered that umami began to be acknowledged as a fundamental taste[3,4].
Umami, as a fundamental taste, is generated through complex physiological processes. Initially, umami substances bind to umami receptors, triggering an allosteric effect and producing gustatins. Subsequently, gustatins are further converted into electrical taste signals. Finally, taste signals are transmitted to the cerebral cortex by nerve fibers and sensed by the gustatory center[5,6]. Umami substances come in various forms, mainly including free amino acids and their salts, nucleotides and their salts, organic acids, organic bases, and umami peptides[7,8]. In recent years, umami peptides have emerged as a new form of umami substance that not only enhances the umami taste but also enriches the nutritional value of food, thus becoming a new research focus. Additionally, umami peptides can synergize with salt to enhance the salty taste and reduce the use of salt for better human health[9−11]. Therefore, it is highly significant to study umami peptides.
Marine organisms have long been recognized as a crucial source of nutrients for humans, particularly high-quality dietary protein[12,13]. Oceans cover 71% of the Earth's surface area and contain vast amounts of resources. In comparison to land-sourced protein, marine protein production causes less environmental stress[14]. Various marine proteins possess a delicious and unique taste and are important raw materials for the preparation of umami peptides[15]. Furthermore, the extraction of umami peptides from marine proteins is an important method to promote the development and utilization of marine resources, which holds significant strategic importance[16−18].
Compared with the land-sourced umami peptides, marine umami peptides have their advantages: Firstly, they are more widely sourced as the area of the ocean is nearly two and a half times larger than that of the land on earth, and the diversity of species in the ocean is far greater than that on land. Secondly, marine umami peptides have higher nutritional value as they usually contain ω-3 polyunsaturated fatty acids (PUFA), minerals, and trace elements such as iodine, zinc, and selenium, which are important for human health. Lastly, marine organisms are rich in umami substances, resulting in an intense umami taste with a more obvious umami-enhancing effect[19−21]. However, it should be noted that current research on marine umami peptides is still in its infancy. The preparation and extraction of these peptides are traditional and lack systematic summary. Therefore, based on existing research findings, summarizing the current progress of marine umami peptides and prospecting new technologies for their application would be beneficial for subsequent research and development efforts in this field.
In this review, the sequence and structural characteristics of reported marine umami peptides are summarized and analyzed systematically. Then, the recognition mechanism of marine umami peptides and their receptors are introduced, followed by the analysis of the signal transduction mechanism. Next, the preparation and regulation techniques of marine umami peptides are summarized. Finally, the application of Industry 4.0 technologies (molecular simulation, artificial intelligence, blockchain, big data, cloud computing) in the study of marine umami peptides are introduced and their future development is prospected. This review aims to introduce the current research of marine umami peptides and prospect the future trends of the methods applied in the subsequent study of marine umami peptides.
-
The amino acid sequence and source information of the reported marine umami peptides that have been reported were shown in Supplemental Table S1 and the distribution of the identified marine umami peptides are shown in Fig. 1. To date, 141 marine umami peptides have been identified, of which 104 are short-chain peptides (dipeptides to nonapeptides), accounting for 73.76%. The number of medium and long-chain polypeptides (decapeptides to eighteen peptides) is 37, accounting for 26.24%. It can be seen that most of the marine umami peptides are short chain peptides. This is consistent with previous studies of umami peptides from other sources[8,22,23]. Specifically, the top three types of marine umami peptides are heptapeptides (23, 16.31%), nonapeptides (17, 12.06%) and hexapeptides (14, 9.93%). The last four types of marine umami peptides are tridecanoic peptides (3, 2.13%), tetradeca peptides (3, 2.13%), hexadecapeptides (2, 1.42%) and octadecapeptides (1, 0.71%). It can be observed from this that the longer the peptide chain, the lower the likelihood of it being umami peptides. In addition, from the view of the amino acid composition of umami peptides, it is found that 85.11% of 120 marine umami peptides contain umami amino acids (glutamic acid or aspartic acid). Zhang et al.[24] summarized the amino acid composition of umami peptides and found that among all umami peptides, those containing umami amino acids accounted for 79.00%, which was lower than the 85.11% found in marine umami peptides. This suggests that marine umami peptides contain a higher proportion of umami amino acids.
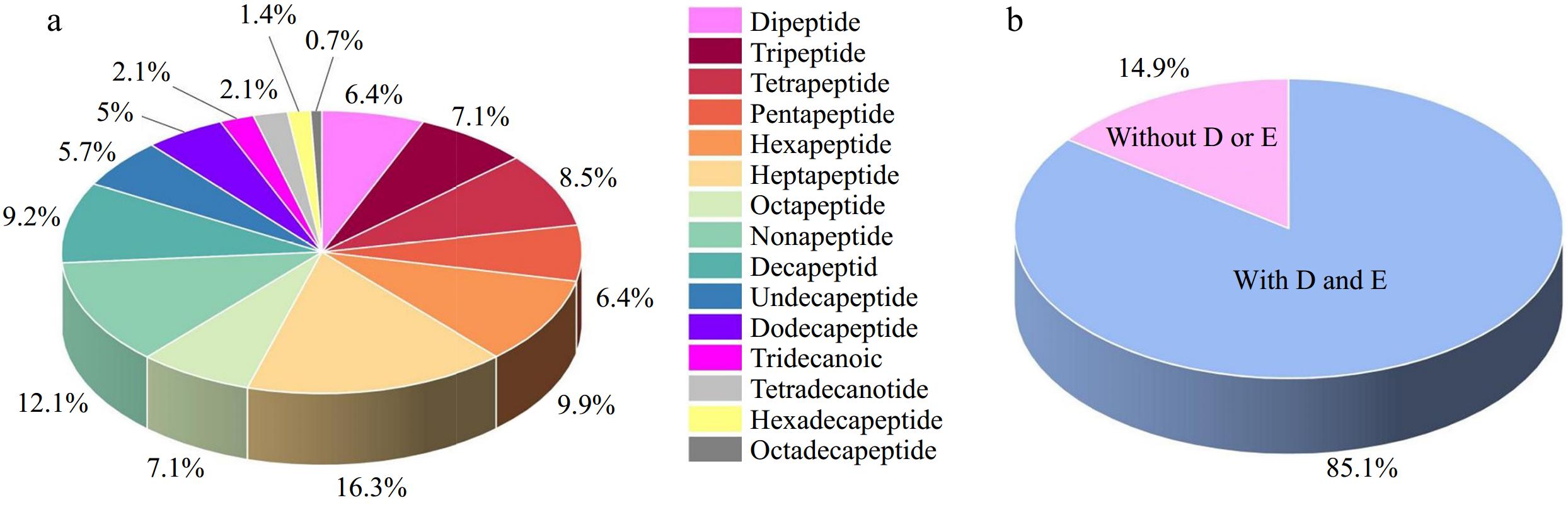
Figure 1.
The information of the identified marine umami peptides. (a) The distribution of the identified marine umami peptides. (b) The percentage of marine umami peptides with umami amino acid (D: aspartic acid, E: glutamic acid).
Furthermore, the types and sequences of amino acids that compose umami peptides also have a great influence on the taste characteristics of peptides[25]. The study of Arai et al.[26] found that the umami taste can only be presented when hydrophilic amino acid is located at the C-terminus. Zhang et al.[24] proposed that the proportion of alkaline amino acids in umami peptides should not be excessively high, as alkaline amino acids are typically found at the end of the polypeptide chain. The amino acid fingerprints of the reported marine umami peptides are shown in Supplemental Fig. S1. In all identified marine peptides, the top four amino acids are glutamic acid (E), accounting for 11.41%, alanine (A) (9.41%), leucine (L) (9.32%) and aspartic acid (D) (9.22%), which are mainly umami/acidic amino acids and hydrophobic amino acids. The last four amino acids are tyrosine (Y) (1.62%), cysteine (C) (1.24%), tryptophan (W) (1.24%). According to the classification of amino acid properties, the proportion of acidic/umami amino acids is 20.63%. The data for alkaline, hydrophilic and hydrophobic amino acids is 13.30%, 29.85%, and 36.23%, respectively (Fig. 2). It can be seen that among all the amino acids composing marine umami peptides, the proportion of acidic/umami amino acid is significantly higher than that of alkaline amino acids, and the proportion of hydrophobic amino acids is higher than that of hydrophilic amino acids. Regarding the order of peptide sequences, it can be observed that hydrophilic and basic amino acids are primarily distributed at the ends of the peptide chain, while hydrophobic and acidic amino acids are mainly distributed in the middle of the peptide chain.
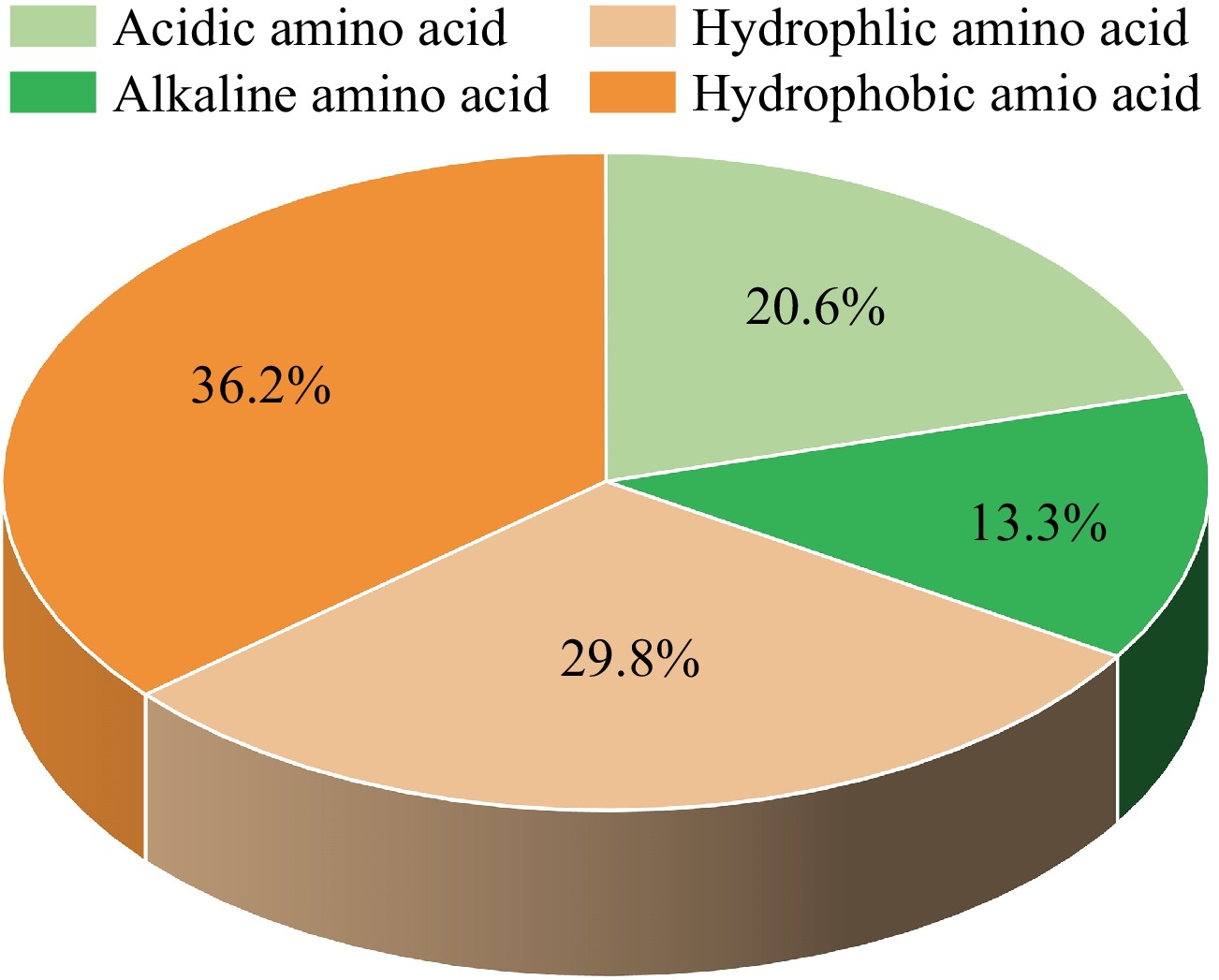
Figure 2.
The amino acid composition of the reported marine umami peptides according to the amino acid features.
The advanced structure of marine umami peptides mainly refers to its secondary structure, including α-helix, β-folding and random curling, etc. The unique marine environment (high salt, low temperature, and high pressure) contributes to the strong stability and adaptability of the structure of marine umami peptides. For instance, the α-helix structure in marine umami peptides enhance their stability, allowing them to maintain biological activity even in extreme environments. In addition, marine umami peptides may also contain special modification groups, such as phosphate groups, sugar groups, etc., which can increase their water solubility and biological activity[27,28].
The taste mechanism of marine umami peptides
-
The taste formation mechanism of marine umami peptides is similar to that of umami peptides from other sources. Initially, umami peptides and receptors (T1R1/T1R3, mGlu1/4) recognize each other in the mouth, leading to the allosteric effects[29,30]. Once the receptor is activated, it produces the corresponding umami gustducin, which further transforms into an umami signal further. Subsequently, the signal molecules bind to their respective receptors and activate taste bud ion channels located on the cell membrane. This activation leads to the release of signal molecules from the cell and their transmission to the nerve center through the taste nerve fibers, resulting in the perception of umami (Fig. 3)[31,32].
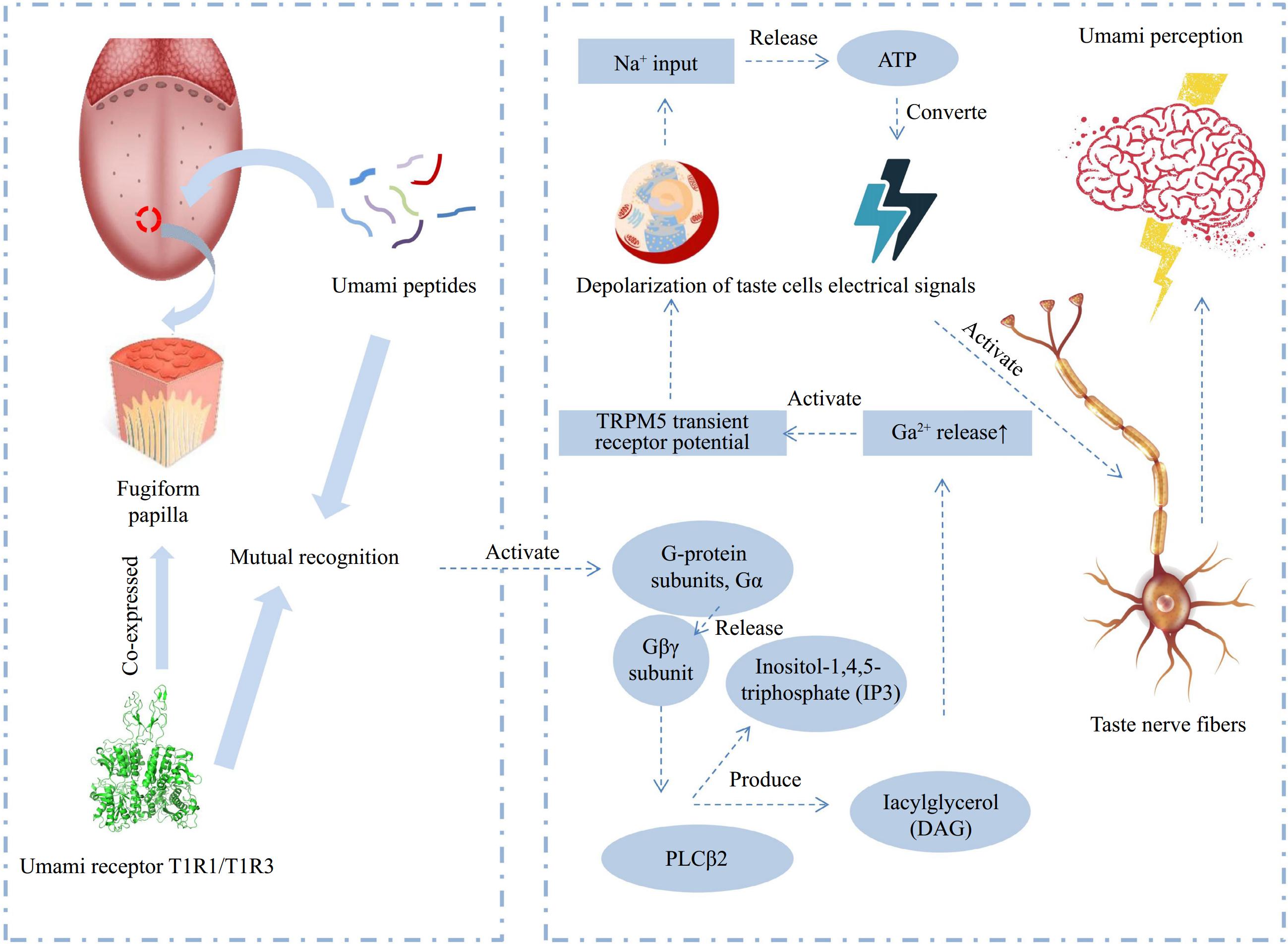
Figure 3.
Schematic diagram of the flavor mechanism of umami peptides (adapted from[32]).
Umami receptors
-
The receptors for umami peptides are a class of membrane proteins called G-protein-coupled receptors (GPCRS). Specifically, the umami peptide receptors mainly include T1R1 and T1R3. T1R1 is co-expressed with T1R3. T1R1/T1R3 heteromer in humans is primarily responsible for detecting and responding to umami substances[29,33,34]. When the umami substance interacts with the T1R1/T1R3 receptor, T1R1/T1R3 initiates signaling pathways that transmit taste information to the brain, resulting in the perception of umami taste[35]. Besides T1R1/T1R3, the umami receptors also include some metabotropic glutamate receptors (mGluRs). MGluRs are also members of the GPCRs family, responsible for the neurotransmitter glutamate. There are eight subtypes of mGluRs, which are classified into three groups based on sequence homology, signal transduction mechanisms, and pharmacological properties. The identified mGluRs umami receptors include mGluR1[36] and mGluR4[37].
Recognition and transduction mechanism of marine umami peptides
-
The analysis of the recognition and transduction mechanism of marine umami peptides is an effective way to study the taste mechanism of umami peptides and mine new umami peptides. Among all umami receptors, heterodimer T1R1/T1R3 receptors are mainly distributed in the front of the tongue. They come into contact with umami substances once the umami substances get into the mouth. Other receptors (mGluR1 and mGluR4) located in the back of the tongue, which results in the lag in its contact with umami substances. Therefore, heterodimer T1R1/T1R3 receptors were identified as the best candidates for umami receptors[38]. All members of the T1R class have a similar structure, that is, the seven-spanning transmembrane region (7TM) and the N-terminal extracellular terminal region[39]. The extracellular terminal region consists of a large 'venus fly trap' (VFT) and a small cysteine-rich domain (CRD). The VFT is the main binding domain for umami ligand recognition, while the CRD mainly connects the VFT domain and the 7TM domain, which is responsible for the transmembrane transmission of the structural changes that occur after VFT ligand recognition[40].
At present, the mechanism of the interaction between umami peptides and umami receptors has not been clarified clearly[32]. Evidence from immunocytochemical and molecular studies suggest that stimulation of umami receptor heterodimers T1R1/T1R3 by umami substances activate the G protein subunit Gα, leading to the release of the Gβγ subunit and stimulating the phospholipase PLCβ2 pathway, which produces inositol-1,4,5-triphosphate (IP3) and diacylglycerol (DAG)[41,42]. The increase of Ca2+ activates the TRPM5 transient receptor potential, leading to the depolarization of taste cells. Taste cells then evoke action potentials through sodium channels and release ATP, which is converted into electrical signals to activate taste nerve fibers, thereby generating umami perception[43]. Although it has been widely recognized that umami receptor heterodimers T1R1/T1R3 stimulate the G protein subunits leading to umami signal transduction, the interaction between umami receptors and ligands and the conformational changes of the receptors after binding umami substances, remain to be examined systematically.
-
Marine umami peptides refer to peptides prepared from marine proteins with an umami taste. The processing methods for these peptides can vary due to the diverse sources of marine proteins. Common processing methods include microbial fermentation, enzymatic hydrolysis, acid hydrolysis, high-temperature processing, and solvent extraction (see Table 1).
Table 1. Overview of the preparation methods for marine umami peptides.
Methods Mechanism Advantages Disadvantages Ref. Microbial fermentation Microbial metabolism degrades marine proteins into peptides Simple operation, safety and low cost High environmental requirements [50,87] Enzymatic hydrolysis Proteases specifically cleave hydrolytic peptide bonds Mild reaction conditions, green environmental protection, and high yield Poor control of the reaction leads to the production of undesirable by-products [21,50,51,54] Acid hydrolysis Proteins are hydrolyzed under acidic conditions Low cost and high yield Toxic substances are easily produced in the reaction process [66,100] High-temperature
processingProtein degrades into small parts under thermal treatment Simple operation and high efficiency High energy consumption and high requirements for equipment [30,68−72] Solvent extraction The majority of umami peptides are soluble in water Simple operation and low cost Low efficiency [44,75] Microbial fermentation
-
Microbial fermentation is a method that utilizes microbial metabolism to degrade marine proteins and obtain marine umami peptides. The vigorous metabolic reaction of microorganisms coverts macromolecular organic matter into amino acids, peptides, organic acids, and other substances[44−46]. Yang et al.[47] found that the production of umami peptides based on microbial metabolism in Chouguiyu was strongly correlated with 22 protease-producing microbial genera, among which Vagococcus, Peptostreptococcus, Acinetobacter, Psychrobacter, and Enterococcus played a major role in the formation of umami peptides. Microbial fermentation has the advantages of fast speed and low cost. At the same time, microbial fermentation does not require the separation and purification of proteins and enzymes, which reduces the production cost and simplifies the operation[48]. Therefore, the use of proteolytic microorganisms to extract umami peptides from animal and plant proteins is more cost-effective and practical than traditional enzymatic hydrolysis. However, microbial fermentation requires high environmental hygiene and is easily contaminated by other bacteria during the fermentation process. As a green and environmentally friendly method for the preparation of marine umami peptides, microbial fermentation has great potential for umami peptide preparation. Future research should focus on the screening of highly resistant strains to achieve more application prospects.
Enzymatic hydrolysis
-
Enzymatic hydrolysis is one of the commonly used methods for the preparation of marine umami peptides. Enzymes can break down proteins into peptides with different molecular weights and free amino acids, which are key umami compounds[49]. Various enzymes such as complex protease[50], pepsin, trypsin[51−54], flavourzyme, papain[21], chymotrypsin[52,53] are commonly employed in enzymatic hydrolysis. The reaction conditions for enzymatic hydrolysis are mild, safe and efficient, easily controlled, and do not produce toxic and harmful substances. This makes it a key technology for developing seafood seasoning[55,56]. Studies have shown that sequential hydrolysis of endonuclease and exonuclease can improve efficiency, reduce bitterness, and enhance the umami taste[57,58]. Flavor protease contains both endo-cut enzyme and exo-cut enzyme activities. These enzymes effectively degrade exposed hydrophobic groups and hydrolyze certain flavor precursor substances to facilitate the formation of high-quality flavor products[59]. To enhance proteolysis and prepare good flavor umami peptides, many researchers have utilized double enzyme hydrolysis methods. For example, Deng et al.[21] prepared and identified umami peptides from Trachinotus ovatus using enzymatic hydrolysis with papain and flavourzyme. Additionally, a stepwise dual-enzymatic hydrolysis process using alkaline and flavor protease was employed to prepare umami enzymatic hydroxylate from squid processing by-products[60].
Acid hydrolysis
-
Under acidic conditions, proteins can undergo hydrolysis reactions to produce peptides or amino acids[61]. Acid hydrolysis is characterized by its simple process, low cost and high-efficiency, making it a widely used method for protein hydrolysis[62−64]. Hydrochloric acid is often used to hydrolyze food proteins to produce umami substances for food flavoring[61,65]. However, during the acid hydrolysis process of preparing marine umami peptides, toxic and carcinogenic substances such as chloropropanol could be generated as by-products[66]. Compared with enzymatic hydrolysis, acid hydrolysis requires more severe reaction conditions[67], which has led to its gradual replacement by other methods.
High-temperature processing
-
Many researchers have utilized high-temperature methods for the preparation of water extracts containing umami peptides[68−72]. During the heating process, proteins degrade into smaller components[73,74]. Wang et al. obtained 29 peptide fragments through high-temperature water boiling combined with gel ultrafiltration and gel filtration chromatography and identified four of them as umami peptides[75]. Additionally, three umami peptides were isolated and purified from Leccinum extremiorientale using high-temperature cooking[76]. Alim et al. separated and purified 15 types of umami peptides from thermally treated yeast extract, with an optimum reaction temperature of 110 °C for umami peptide generation[77]. High-temperature processing is a common method for extracting nutrients from foods; therefore, the process of preparing umami peptides by this method is simple and suitable for industrial production. However, it is important to strictly control the temperature and heating time to avoid excessive hydrolysis and denaturation of the protein.
Solvent extraction
-
Because most umami peptides are water-soluble, water-based solvent extraction is commonly used for their extraction. Hot water extraction is a traditional method for obtaining umami peptides. Four umami peptides were identified from Lactarius volemus aqueous extracts, with one peptide (EVAEALDAPKTT) having a very low umami threshold of only 0.0625 mg/mL[78]. To enhance extraction efficiency, solvent extraction is often combined with high-temperature treatment, homogenization, ultrasonic treatment, and other methods[61]. Wang et al. extracted proteins and peptides from Takifugu flavidus using homogenization and a water-based solvent extraction method, resulting in the identification of four peptides with umami taste[79]. The process of solvent extraction is straightforward and does not require expensive equipment. However, it has the disadvantage of high energy consumption and long extraction time[61]. In recent years, there has been a growing interest in natural green extraction solvents, such as aqueous two-phase and deep eutectic solvent (DES), for extracting bioactive substances, making it a research hotspot[80,81]. These solvents have also been applied to the extraction of marine umami peptides. For example, DES composed of glucose, sucrose, and water was used to enhance the umami-enhancing capacity of pea protein hydrolysate[82]. Although there are currently few reports on the application of natural green extraction solvents, their potential in the extraction of marine umami peptides can be seen based on their extensive use in extracting bioactive substances[83−86].
Regulation techniques of umami taste
-
During the process of preparing marine umami protein and peptides products, it is inevitable that some unique substances from marine organisms will be incorporated into them, such as ω-3PUFA and astaxanthin. However, these substances will produce many unpleasant flavors after oxidation. Moreover, umami peptides may be decomposed by temperature, pH, and other conditions during processing or storage, leading to a significant reduction in their umami taste. Therefore, it is necessary to use embedding and adding amino acids in the actual production process to regulate the umami taste and maintain the stability of these products.
Embedding methods
-
Embedding is a technique used to immobilize active substances (such as drugs, enzymes, proteins, etc.) in a carrier material. This technique serves various purposes including protecting the active substance, prolonging its duration of action, improving stability, and controlling the release rate[87]. In the food industry, embedding can also be utilized to immobilize food additives or condiments in carriers to achieve specific release characteristics. Currently, spray drying and freeze drying are commonly employed methods for embedding umami peptides[88]. The process of microencapsulation through spray drying and freeze drying involves preparing a mixed emulsion of heartwood and wall material first, followed by spraying or freeze-drying the emulsion in a desiccator[89]. While these methods effectively prevent the oxidation of umami peptides and their reaction with other substances which could weaken their flavor profile, they do have drawbacks such as high energy consumption and low automation control level[90].
Amino acid addition
-
The amino acid addition method is a technique to regulate the umami taste by adding pure amino acids or amino acids mixture into the umami protein or peptide products. These amino acids or amino acids mixtures can cooperate with other flavorful peptides, nucleotides, and other substances contained in food to further enhance umami taste. Fu et al.[91] improved the umami taste of the umami peptides in fish head soup by adding cysteine. Similarly, Ruan et al.[92] maintained the umami taste of soy sauce by adding hydrolysate from low-value fish. The addition of amino acids is a direct method for increasing the umami flavor of products. While this process is simple, it requires exploration and optimization of the amount of added amino acids in the early laboratory stages, which may also result in additional economic costs being incurred at that time.
-
Recently, various global challenges, including global warming, increasing global population, overfishing, and other ecosystem damage, make it necessary for human beings to further innovate, study, and carry out sustainable development and utilization of marine resources. Therefore, Industry 4.0 technology has received great attention in recent years to improve efficiency and productivity and enhance sustainability[93,94]. Industry 4.0, categorized according to different stages of industrial development represents an era in which information technology is utilized to drive transformation within the industry. The artificial intelligence, big data analysis, Internet of Things, blockchain, smart sensors, robotic and cyber-physical systems are some of the technologies featured in Industry 4.0[95,96], and emerges as a way to improve the competitiveness of marine food[97,98]. Industry 4.0 technology has been used widely in the simulation of the interaction between the marine umami peptides and their receptors, the rapid screening and structure prediction of marine umami peptides, and the establishment and sharing of marine umami peptides database[99−101]. Furthermore, Industry 4.0 technology holds significant potential for the collection and processing of characteristic data and traceability analysis of marine umami peptides. This section introduces and prospects the application of Industry 4.0 technologies (molecular simulation, artificial intelligence, big data, cloud computing, and blockchain) in the study of marine umami peptides (Fig. 4), aiming to promote their application in the subsequent scientific research of marine umami proteins and peptides.
Molecular simulation
-
Molecular simulation (MS) technology is a method that uses computers to simulate the behavior and properties of molecular systems at the atomic or molecular level[102]. MS technology has been applied to the study of marine umami peptides, mainly including the following aspects: (1) Interaction simulation: the interaction between marine umami peptides and other molecules (such as receptors and enzymes) can be simulated, and the mechanism of action and binding mode in vivo can be predicted, which is helpful for studying the biological activity and pharmacological effects of marine umami peptides. The interaction between shrimp umami peptide and its receptor was simulated through homology modeling and molecular docking and the results indicated that amino acid residues Arg151, Asp147 and Gln52 might be the key binding sites[103]. Five umami peptides were obtained from Meretrix lusoria, and explored the interaction between these umami peptides with T1R1/T1R3 by molecular simulation technology[19]. The results indicated that the peptides could enter the binding pocket in the Venus flytrap domain of the T1R3 cavity, wherein Asp196 and Glu128 may play key roles in the sensation of umami taste. Moreover, hydrogen bonding and electrostatic interactions are important interaction forces. (2) Structural prediction: molecular simulation technology can help researchers to predict and simulate the molecular structure of marine umami peptides, including three-dimensional structure, conformation and stability analysis of proteins, peptides, and other molecules, so as to help understand their functions and biological activities. Bu et al.[87] studied the characterization and structure-activity relationship of novel umami peptides isolated from Thai fish sauce by molecular simulation technology combined with quantitative structure–activity relationship (QSAR). The results indicated that the umami peptide chains were rotated and folded obviously, and hydroxyphenyl group of the N-terminal tyrosine residue side chain rotated significantly, which may enhance the umami taste. Although molecular simulation technology can effectively simulate the structural change process of biological macromolecules dynamically, the internal structure of biological systems, with uncertainties that change over time, is much more complex than imagined[104]. Therefore, the simulation process of biomacromolecules also has certain limitations, which require the improvement of computing software and hardware as much as possible to meet the needs of solving practical problems.
Artificial intelligence
-
Artificial intelligence (AI) is the technology of creating and applying intelligent machines or software that can mimic, augment, and extend human intelligence. The research field of artificial intelligence covers robotics, language recognition, image recognition, natural language processing, etc. The goal is to make computers have the ability to think and make decisions like humans[105,106]. The application of AI in marine umami peptides mainly includes the following aspects: (1) Discovering new marine umami peptides involves the use of advanced techniques such as machine learning and deep learning. By analyzing large quantities of known peptides, researchers can model and predict their structure and taste. This approach holds great potential for advancing our understanding of marine umami peptides and their applications in various fields. Meanwhile, virtual screening combined with AI could be used to determine their biological activities and taste characteristics, helping to accelerate the discovery of new marine peptides. Zhang et al.[107] investigated an interpretable BERT-based AI model to realize rapid screening of new umami peptides with a computational accuracy of 93.23%. Qi et al.[108] developed a novel peptide sequence-based umami peptide predictor, namely Umami-MRNN, which was based on multi-layer perceptron and recurrent neural network. The independent tests have shown that Umami-MRNN achieved an accuracy of 90.5%. (2) Prediction of allosteric effects can be achieved by integrating and analyzing a substantial amount of experimental data, in combination with methodologies such as machine learning and network analysis. This approach allows for the elucidation of the mechanism of action and signaling pathways associated with marine umami peptides. This helps to understand the function of marine umami peptides in cells and provides a theoretical basis for further application and development. Cui et al.[109] predicted the conserved sites and recognition mechanisms of T1R1 through ensemble docking combined with machine learning. The results indicated that residues 107S-109S, 148S-154T, and 247F-249A mainly form hydrogen bonding contacts, which might be the key binding sites during the sensation of umami. The binding processing of two umami peptides (MTLERPW and MNLHLSF) with T1R1 was predicated by Li et al.[110] through phage display combined with AI, and the key binding sites (MW-7 and MF-7) in the VFT were identified. (3) Utilizing AI technology, the analysis of large-scale marine umami peptide sequences can be conducted through bioinformatics techniques such as sequence alignment, structure prediction, and functional annotation. This approach is beneficial for predicting the characteristics of marine umami peptides and establishing a reference threshold for their properties. An AI system for taste analysis based on a graph neural network was developed by Lee et al.[33], which could predict the taste threshold of marine umami peptides based on their structure, providing a new method for the perception research of marine umami peptides. AI relies on a large amount of high-quality data for training and verification, with the database serving as the foundation for AI model testing and training[111]. AI relies on a large amount of high-quality data for training and verification, with the database serving as the foundation for AI model testing and training[111]. Therefore, AI requires a substantial volume of accurate data and computational resources to train, optimize, and support research related to umami peptides. The collection of such data has become a limiting factor in the application of AI in the development of umami peptides. However, as the model improves and more data is accumulated, AI will create new opportunities for the advancement of marine umami peptides.
Big data
-
Big data technology is a data processing technique that utilizes computer systems and network technology to process massive amounts of data. It aims to extract useful information from the data through association analysis and prediction. Big data technology is characterized by its volume, velocity, variety, and value. The primary function of big data is to record, describe, and predict various phenomena[112−114]. In the context of marine umami peptides, the application of big data encompasses several key aspects. Firstly, it involves the establishment and sharing of databases. By integrating and sharing characteristic data related to marine umami peptides, a specialized database can be developed. This database is a valuable resource for studying marine umami peptides, ultimately expediting research processes and fostering collaboration within the field. Gradinaru et al.[115] and Rojas et al.[116] collected a large number of characteristic information of umami substances through big data technology and established two open access umami databases (PlantMolecularTasteDB and ChemTastesDB), which greatly facilitated the subsequent research on marine umami peptides. Secondly, by utilizing big data analytics, marine biological scientists and researchers can effectively process large-scale marine biological data to gain a better understanding of the genomic, proteomic, and metabolomic features of marine organisms. This will enable them to explore the biosynthetic pathways and properties of umami peptides[117]. Finally, the promotion and marketing of marine umami peptide products can benefit from the application of big data technology for consumer behavior analysis. By analyzing consumer data and market trends, a better understanding of consumer needs and preferences can be obtained, which in turn can guide the promotion and marketing strategies of marine umami peptide products. This approach allows for a more targeted and effective marketing strategy that is based on empirical evidence and insights into consumer behavior[118]. From a functional perspective, the application of big data in marine umami peptides primarily involves the collection of various relevant data. Therefore, the challenges of applying big data in marine umami peptides lie in the capacity, source, and quality of the data[119].
Cloud computing
-
Cloud computing technology is an internet-based computing model, which provides a variety of computing services through the network, including storage, database, software, analysis and processing capabilities. The application of cloud computing in the development and utilization of marine umami peptides are still in their early stages. The application might include the following aspects: (1) Data storage and processing: the study of marine umami peptides requires a large amount of data storage and processing, including gene data and protein data of marine biological samples. Cloud computing can provide efficient data storage and processing capabilities to help researchers quickly analyze the components and characteristics of marine umami peptides. (2) Virtual experiments and simulations: through the cloud computing platform, researchers can conduct virtual experiments and simulations to explore the biological activities, pharmacological effects and other characteristics of marine umami peptides, to accelerate research progress. Although the function of cloud computing has greatly facilitated the discovery of new marine umami peptides, the nature of cloud computing also brings challenges for cloud computing applications[120,121]. The main challenges of cloud computing in marine umami peptide is data security[19]. Because the cloud computing provider has complete control over all operations, malicious detection, service manipulation and economic denial-of-service attacks by the provider are the potential threats of data being damaged or breached[121].
Blockchain
-
Blockchain technology is a distributed database-based technology that is capable of recording and storing transaction data. It possesses the characteristics of decentralization, non-tampering, and transparency. At its core, blockchain technology consists of a chain structure composed of a series of blocks, with each block containing a specific amount of transaction data. The security and consistency of the data are ensured through encryption algorithms and consensus mechanisms[122]. Blockchain technology has a wide range of applications in the downstream management of marine umami peptides. Firstly, through the use of blockchain technology, it is possible to achieve full traceability of marine umami peptide products. This means that every stage in the production, processing, and transportation of each batch of products can be recorded on the blockchain. As a result, consumers can access detailed information about the product such as its source, production date, and production process by scanning the two-dimensional code on the product or querying relevant information. This increased transparency and credibility serves to enhance consumer confidence in the product[123]. Secondly, the authenticity of marine umami peptide products can be verified using blockchain technology. Each batch of products is assigned a unique blockchain identity, allowing consumers to verify the product's authenticity by scanning the two-dimensional code or entering relevant information. This ensures that the purchased product is genuine[124]. Thirdly, supply chain management: Blockchain technology has the potential to enhance the supply chain management of marine umami peptide products[125]. By integrating all aspects of the supply chain into a blockchain network, real-time monitoring and transparent management can be achieved. This reduces information asymmetry and risk within the supply chain while improving efficiency and reliability[126]. The high-cost problem is a significant challenge when applying blockchain technology in the marine umami industry. Embedding blockchain technology into the marine umami peptide traceability system requires a substantial investment of both time and money for participants. As the complexity of the blockchain increases, so does the need for additional computing power to confirm more blocks, resulting in higher power consumption[127]. Furthermore, as the number of transactions within the marine umami peptide tracking system grows, so does the volume of data. This increase in data creates challenges related to storage and computation, ultimately reducing the capacity scale of the system and increasing synchronization time for new users[127,128].
-
As a treasure house of natural resources, the ocean contains a huge amount of marine proteins. The preparation of umami peptides from these marine proteins is an important way to utilize ocean resources. This review summarized the amino acid composition, characteristics, and sequence information of the reported 141 marine umami peptides. Additionally, the taste mechanism and preparation methods of marine umami peptides were introduced. According to the current research results, most marine peptides are composed of four to ten amino acids and abundant in umami/hydrophobic amino acids. The preparation methods of marine umami peptides include microbial fermentation, enzymatic hydrolysis, acid hydrolysis, high-temperature processing, and solvent extraction. Each method has its advantages and disadvantages, better results may be obtained by using the combined methods. At the same time, embedding methods and amino acid addition were commonly used to adjust umami taste, which had the advantages of simple operation and was suitable for industrial production. Finally, the existing ongoing projects regarding the application of Industry 4.0 technology in marine umami peptides were introduced and the future trends of its application prospected. Based on the current state of research, several Industry 4.0 technologies (molecular simulation, artificial intelligence, and big data) have been widely utilized in simulating the interaction between the marine umami peptides and their receptors, rapidly screening and predicting the structure of marine umami peptides, as well as establishing and sharing a database of marine umami peptides. The use of cloud computing and blockchain is still in its early stages. Prospective applications may include the collection and processing of characteristic data, traceability analysis of marine umami peptides etc. Essentially, the core technology of Industry 4.0 primarily serves as information storage and data processing tools that greatly enhance researchers' work efficiency. Furthermore, in order to better understand the properties and applied research of marine umami peptides, it is necessary to encourage and stimulate more investment in biotechnology to promote the sustainable development and utilization of these marine resources in the future.
-
The authors confirm their contribution to the paper as follows: data curation: Hu D, Zheng Z, Liang B, Jin Y, Shi C, Chen Q, Wei L; writing-original draft: Hu D, Zheng Z; writing-review & editing: Dong X, Lu Y; conceptualization: Jin Y, Shi C, Lu Y; literature arrangement: Chen Q, Wei L; software, investigation: Li D, Li C; project administration, resources: Ye J, Dai Z; funding acquisition: Lu Y. All authors reviewed the results and approved the final version of the manuscript.
-
All data generated or analyzed during this study are included in this published article.
The authors are grateful to the National Key Research and Development Program of China (2023YFD2100203), the Natural Science Fund of Zhejiang Province (LQ22C200008) and Basic research funds for provincial colleges and universities (FR2401ZD).
-
The authors declare that they have no conflict of interest.
-
Authors contributed equally: Di Hu, Zhenxiao Zheng
- Supplemental Table S1 The amino acid sequence and source information of the reported marine umami peptides.
- Supplemental Fig. S1 The amino acid fingerprints of the reported marine umami peptides (a) dipeptide ~ hexapeptide; (b) hexapeptide ~ octapeptide; (c) octapeptide ~ dodecapeptide; (d) tridecanoicpeptide ~ octadecapeptide.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press on behalf of China Agricultural University, Zhejiang University and Shenyang Agricultural University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Hu D, Zheng Z, Liang B, Jin Y, Shi C, et al. 2024. Recent progress and prospects in production and identification of umami peptides from marine proteins. Food Innovation and Advances 3(3): 256−267 doi: 10.48130/fia-0024-0024
Recent progress and prospects in production and identification of umami peptides from marine proteins
- Received: 14 May 2024
- Revised: 08 July 2024
- Accepted: 12 July 2024
- Published online: 09 August 2024
Abstract: Umami peptides, the flavor compounds mainly derived from natural proteins, provide a pleasant taste for humans and exhibit a variety of biological activities, such as antioxidant and lipid-lowering properties. Marine proteins, which serve as excellent sources of umami peptides, have become a focal point of research. This review introduces the research progress on reported marine umami peptides. Firstly, it discusses the structural characteristics of umami peptides and the mechanism behind their formation to create an umami taste. It then presents several commonly used techniques for preparing and regulating umami peptides while summarizing the advantages and disadvantages of each technique. Finally, this review describes the potential application prospects for core technologies within Industry 4.0—such as molecular simulation, artificial intelligence, big data analysis, cloud computing, and blockchain technology—which could bring new opportunities for the development of marine umami peptides.














