-
Transient gene expression is a powerful and versatile method for quickly assessing gene expression in higher plants. This technique, particularly when mediated by Agrobacterium, involves the transcription of non-integrated T-DNA genes within plant cells[1], and it has been widely applied in various fields including the production of recombinant protein, subcellular protein localization, protein-gene interaction studies, functional genomics, and epigenetic regulation assays[2−6]. In citrus, leaf tissues have been the preferred choice for Agrobacterium infiltration due to their ease of handling and high transformation efficiency[7]. However, other tissues, including citrus fruits[8], and phloem-enriched organs such as immature and mature shoots are increasingly being used for specialized applications[9].
One critical application of Agrobacterium-mediated transient gene expression in citrus is the study of Huanglongbing (HLB), a devastating bacterial disease that targets the phloem of citrus plants. HLB, as a result of Candidatus Liberibacter asiacticus (CLas) infection causes the accumulation of callose in phloem sieve plates, impairing the transport of photosynthetic products and leading to widespread cellular damage[10]. Research has shown that CLas infection triggers the upregulation of genes associated with ROS production and the downregulation of antioxidant enzymes in the phloem, resulting in the systemic death of phloem cells[11]. Therefore, developing an efficient transient gene expression system using phloem-enriched tissues, such as epicotyls and shoots, is crucial for studying the genetic responses to CLas and other phloem-related phenomena.
The efficiency of Agrobacterium-mediated transient gene expression is influenced by several factors, including the strain of Agrobacterium used and the physiological condition of the plant tissues[12]. Additionally, the composition of the culture media for both Agrobacterium and the plant tissues plays a significant role in determining the success of transient gene expression, as evidenced in various plant species[13−15]. Numerous strategies have been developed to enhance Agrobacterium-mediated transient gene expression, such as optimizing culture conditions and refining the vectors for gene delivery[7,16−19].
The recent advancements in CRISPR/Cas9 and other gene editing technologies have revolutionized the genetic improvement of citrus[20]. Unlike traditional transgenics, gene-edited citrus plants are anticipated to be more acceptable to the public and may face fewer regulatory obstacles, provided they do not contain foreign DNA[21−23]. One method to achieve non-transgenic gene editing is through the delivery of pre-assembled Cas9-gRNA complexes into protoplasts, a technique that has shown promise in various plant species including Arabidopsis, tobacco, lettuce, and rice[24]. However, producing non-transgenic, gene-edited citrus plants remains challenging. Our previous study using Agrobacterium-mediated transient expression demonstrated the potential for creating non-transgenic, gene-edited tobacco plants with high efficiency[25]. Yet, applying this technique to citrus has proven less effective (unpublished data). Therefore, optimizing the transient expression of Cas9 and gRNAs is essential for improving the efficiency of Agrobacterium-mediated gene editing in citrus.
Epicotyls are particularly advantageous for gene editing in citrus due to their high susceptibility to Agrobacterium infection and their capacity for effective shoot regeneration[9,26]. Several research initiatives have successfully employed Agrobacterium-mediated stable expression of Cas9 and gRNA in epicotyl explants to generate gene-edited citrus plants[9,27,28]. Recent work by Alquézar et al. has demonstrated a novel approach for editing the citrus acetolactate synthase (ALS) gene using Agrobacterium-infected epicotyl tissues without the need for antibiotic selection[29]. Additionally, mature citrus stem tissues provide an alternative explant for gene editing, allowing for the evaluation of fruit without the extended juvenile period typical of citrus[30−32]. However, the efficiency of Agrobacterium infection and subsequent shoot regeneration remains a limiting factor when using mature stem tissues.
In this manuscript, an optimized method for utilizing epicotyl tissues of juvenile seedlings and stem tissues of mature citrus as explants for Agrobacterium-mediated transient gene expression is described. By refining various factors, particularly the compositions of the culture media used for Agrobacterium and explant culture, as well as for Agrobacterium-explant co-cultivation, up to a 6-fold increase in transient gene expression activities was achieved in the epicotyls of juvenile Carrizo citrange seedlings and stem segments of mature Pineapple sweet orange, Valencia orange, and Washington navel orange trees. This enhanced epicotyl- and mature stem-based transient gene expression system offers significant potential for producing transgene-free, gene-edited citrus plants and for advancing our understanding of genes involved in CLas-plant interactions[33,34].
-
Epicotyl tissues used in this study were sourced from Carrizo citrange seedlings [Citrus sinensis (L.) Osbeck × Poncirus trifoliata (L.) Raf.]. Seeds were purchased from Tree Source Citrus Nursery (504 N Kaweah Ave, Exeter, CA 93221, USA). Seeds were manually de-coated and then surface-sterilized by treating with 75% ethanol for 60 s, followed by a 20-min soak in 1% sodium hypochlorite, and finally rinsing 4 times with sterile distilled water. Subsequently, internal seed coats were removed, and the seeds were germinated on Murashige and Skoog (MS) medium containing 30 g/L sucrose and 7 g/L agar at pH 5.8. The seeds were kept at 28 °C under various light/dark conditions depending on the specific experimental treatments. Etiolated seedlings were grown in the dark for varying periods, ranging from 3 to 4 weeks. In contrast, light-grown seedlings were initially germinated in the dark for 3 weeks and then transferred to a 16/8-h light-dark cycle with a photon flux density (PPFD) of 60 μmol/m²/s for 1 week or until the etiolated seedlings turned green. To investigate the effect of hormone treatments on transient gene expression, 1-cm-long epicotyl segments were incubated in a liquid MS medium containing 3 mg/L 6-BA, 1 mg/L 2,4-D, and 0.1 mg/L NAA at pH 5.8 for periods of 0, 3, 6, or 9 h before Agrobacterium infection.
Somatic stem tissues were obtained from young growing shoots of newly grafted Pineapple sweet orange [Citrus sinensis (L.) Osbeck], Valencia orange [Citrus sinensis (L.) Osbeck], and Washington Navel orange [Citrus sinensis (L.) Osbeck] trees. These trees were propagated in a greenhouse (maintained at 18−27 °C) by grafting buds from adult mother trees onto 1-year-old sour orange rootstocks, which were grown for approximately 5 months before transformation. The newly growing shoots, approximately 20 cm in length, were harvested, and leaves and thorns were removed. The bud sticks were disinfected by immersion in 1 M HCl for 30 s, followed by treatment with a 20% (v/v) commercial bleach solution containing 0.2% (v/v) Tween 20 for 30 min[30,35]. The bud sticks were then rinsed 5 times with sterile distilled water, and the internodal stems were cut into thin segments for Agrobacterium infection.
Agrobacterium preparation and treatments
-
Agrobacterium tumefaciens strain EHA105, carrying the binary vector pCambia1305.1 with the 35S::GUS fusion gene, was used to evaluate transient gene expression. The Agrobacterium stock was streaked on a solid LB medium plate containing 100 mg/L kanamycin and 50 mg/L rifampicin and cultured at 28 °C for 2 d. Single colonies were transferred to 5 mL of liquid LB medium supplemented with 100 mg/L kanamycin and 50 mg/L rifampicin and cultured at 28 °C with shaking at 200 rpm for 24 h. The 5 mL of cultured bacterial solution was then transferred into 50 mL of liquid LB medium containing the same antibiotics and cultured under the same conditions until the OD600 reached 0.6. Agrobacterium cells were harvested by centrifugation at 5,000 rpm for 15 min and resuspended in a liquid co-cultivation medium comprising MS medium, 30 g/L sucrose, and 20 mg/L acetosyringone (AS), unless otherwise stated.
The effects of AS, different media compositions, and Agrobacterium cells pretreatment incubation times on transient expression were evaluated. Specifically, 20 mg/L AS was added to the LB medium, and Agrobacterium cells were pre-incubated in media containing MS,1/10 MS, ½ MS & ½ AB, and AB for 3, 6, and 9 h before infection, respectively.
Agrobacterium infection and citrus shoot regeneration
-
On the day of Agrobacterium infection, juvenile Carrizo citrange epicotyl seedlings were cut into 1 cm segments under sterile conditions and incubated in the Agrobacterium suspension as described by Orbović et al.[31]. The explants were then blotted dry on sterilized filter paper and arranged end-to-end in Petri dishes containing co-cultivation medium (MS medium, 3 mg/L 6-BA, 30 g/L sucrose, and 20 mg/L AS) and incubated in the dark at 25 °C for 3 d unless otherwise specified. For the mature citrus cultivars, stem segments from the first flushes of grafted citrus plants were used. The mature tissue explants were prepared similarly, with thin segments incubated in the Agrobacterium suspension, blotted dry on sterilized filter paper, and placed in Petri dishes with co-cultivation medium (MS basal medium, 2 mg/L 2,4-D, 2 mg/L IAA, 1 mg/L 2-iP, 30 g/L sucrose, and 20 mg/L AS) and incubated in the dark at 25 °C for 3 d unless otherwise specified.
To identify the optimal co-cultivation duration for maximum transient expression, explants were kept on co-cultivation medium for 2 to 6 d before being transferred to shoot regeneration medium supplemented with 150 mg/L timentin and 20 mg/L hygromycin. They were then cultured under light conditions (60 μmol/m2/s) with a 16-h photoperiod at 26 ± 2 °C for 5 d.
For citrus shoot regeneration, juvenile citrus explants were transferred to a shoot regeneration medium containing MS, 3 mg/L 6-BA, 30 g/L sucrose, and 8 g/L agar and cultured under light conditions (60 μmol/m²/s) with a 16-h photoperiod at 26 ± 2 °C, and they were subcultured onto fresh media every 3 weeks.
Chemical inclusions in co-cultivation media
-
The effect of chemical treatments on transient expression was assessed using media containing various concentrations of sulfamethazine (SMZ) (10 μM, 30 μM, and 50 μM), lipoic acid (LA) (5 μM, 10 μM, and 20 μM), and paclobutrazol (PBZ) (10 μM, 30 μM, and 50 μM), either alone or in combinations, in the co-cultivation media. The incubation treatment lasted for 3 d.
Transient GUS expression analysis
-
The Agrobacterium-infected tissues were stained with a solution containing 100 mM potassium phosphate buffer, 10 mM Na2EDTA, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 0.1% Triton X-100, and 1 g/L X-gluc (5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid) at 37 °C for 12 h. The stained tissues were then sequentially destained in 95% and 70% ethanol until chlorophylls were removed or the tissues became clear. Light microscopy images of the stained tissues were analyzed using ImageJ software to quantify the blue staining intensity (stained area multiplied by color intensity) of each explant[36]. A total of 200 explants were analyzed per treatment.
Data analysis
-
SPSS software was employed for statistical analysis. The results are presented as the mean ± standard deviation (SD) of at least three replicate measurements. To assess significant differences, one-way ANOVA was used. For multiple comparisons, LSD was applied to compare significant differences (p < 0.05) among the different treatments.
-
Agrobacterium cells harboring a Ti-plasmid with a 35S::GUS gene were used to infect the epicotyls of Carrizo citrange, and the infected tissues were histochemically stained for GUS activity. Figure 1a shows that GUS expression appeared as early as the second day post-infection (dpi) in the inoculated epicotyl explants and peaked between 3 dpi and 4 dpi, and gradually declining thereafter. When the infected tissues were transferred to a selection medium for an additional 5 d, GUS expression drastically decreased (Fig. 1b). This expression pattern supports transient GUS expression in the infected tissues at an early stage. However, GUS activity was still observed in tissues on day 11 (6 + 5) (Fig. 1b), although much reduced than on day 3 (Fig. 1a), suggesting that these GUS spots are a result of stably integrated T-DNAs. Therefore, transient GUS expression peaks on the 3rd day after co-cultivation, and the subsequent experiments will be conducted over a 3-d incubation period.

Figure 1.
Agrobacterium-mediated transient GUS expression in the citrus epicotyl explants. (a) Histochemical staining of GUS activity in dark-grown citrus epicotyl explants at 2, 3, 4, 5, and 6 d post Agrobacterium infection (dpi). The blue color indicates the transient and stable expression of the GUS reporter gene. (b) The explants infected with Agrobacterium in the same way as in (a) were transferred to a selection medium containing timentin and hygromycin for an additional 5 d. The differences in GUS activity (blue color) between (a) and (b) are indicative of Agrobacterium-mediated transient expression of the GUS gene.
Growth condition and age of seedlings affect transient GUS expression
-
The effect of growth conditions and seedling age on transient gene expression were next investigated. As shown in Fig. 2a, the dark-grown seedling tissues (two epicotyl explants on the right) exhibited significantly higher transient expression activity compared to the light-grown seedling tissues (2 epicotyl explants on the left). This observation was further confirmed by the quantitative analysis of GUS activities (Fig. 2b). The efficiency of stable transgenic plant production was also assessed using light-grown and etiolated seedlings as explants (Fig. 2c & d) and observed no significant differences between these explants. Additionally, the effect of seedling age on transient GUS expression was examined. Among the 5 stages analyzed (Fig. 2e), 4-week-old and 5-week-old seedlings showed significantly higher levels of GUS expression, whereas 7-week-old seedlings exhibited the lowest levels. In subsequent experiments, 4-week-old etiolated seedlings were used to investigate additional factors influencing transient expression of T-DNA genes.
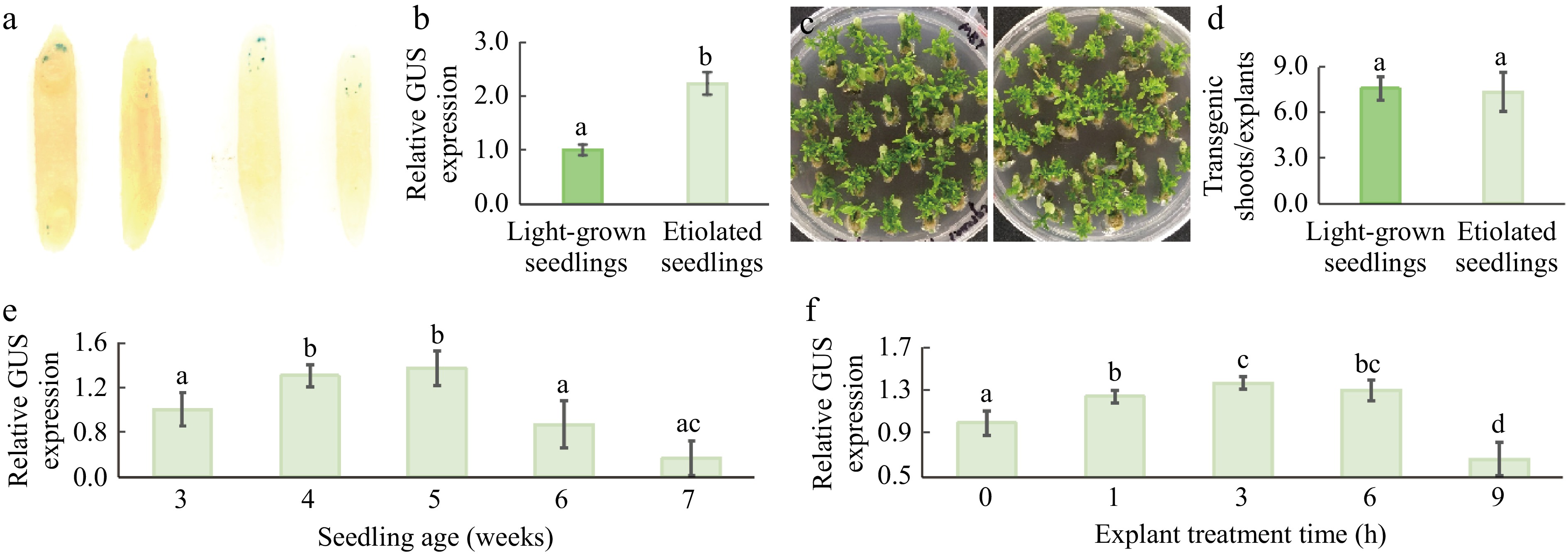
Figure 2.
Effects of growth conditions and seedling age on transient GUS expression. (a) Differential GUS expression in the infected 4-week-old etiolated (two epicotyl explants on the right) and light-grown (two epicotyl explants on the left) tissues. (b) Quantitative analysis of GUS activity in the infected 4-week-old etiolated and light-grown tissues. (c) Transgenic shoot regeneration from the infected etiolated (right panel) and light-grown (left panel) epicotyl explants. (d) Transgenic shoot regeneration efficiency of the infected etiolated and light-grown tissues. (e) Effects of seedling age on GUS gene expression. (f) Effects of duration of hormone treatments of explants on transient GUS expression. The explants were treated in a medium containing 3 mg/L 6-BA, 1 mg/L 2,4-D, and 0.1 mg/L NAA for 0, 1, 3, 6, and 9 h prior to Agrobacterium infection, respectively. Data were averaged from three independent transformation replicates, with ± SD. Significance among treatments is indicated by different lowercase letters (p < 0.05, ANOVA/LSD).
Effect of hormone treatments of etiolated explants before infection on transient gene expression
-
Our earlier study demonstrated that pre-treatment of epicotyl explants with 6-BA, 2,4-D, and NAA enhanced stable transformation efficiency[37]. To determine if the same treatment could also promote transient gene expression, 4-week-old etiolated explants were pre-incubated with a medium containing MS, 3 mg/L 6-BA, 1 mg/L 2,4-D, and 0.1 mg/L NAA[37,38] for 0, 1, 3, 6, and 9 h before infection with Agrobacterium. Figure 2f shows that GUS expression increased with incubation time and peaked between 3 and 6 h before declining. This suggests that hormone treatments of etiolated explants with 3 mg/L 6-BA, 1 mg/L 2,4-D, and 0.1 mg/L NAA for no more than 6 h can enhance Agrobacterium-mediated transient expression of T-DNA genes in citrus epicotyl tissues.
Effects of acetosyringone and co-cultivation medium of transient GUS expression
-
Acetosyringone (AS) has been widely documented to enhance transient expression and stable transformation in various plants[39,40]. To test whether AS could also enhance GUS transient expression in citrus epicotyl tissues, 20 mg/L AS was added to Agrobacterium-growing LB medium and incubated for 8 h before infection of the epicotyl tissues. Figure 3a shows that GUS expression remains invariant between treatments with or without AS, suggesting that AS in LB medium barely affects GUS expression. Given that the maximal induction of expression of vir genes occurs at acid pH 5.2−6.0, pH 7.0 in LB medium could explain why AS fails to stimulate GUS expression in the infected epicotyl tissues. To address this, Agrobacterium cells were resuspended and incubated in various AS-containing basal media with pH 5.8 for 3 h before infection, including MS medium, 1/10 MS, ½ MS & ½ AB (Agrobacterium minimal medium) medium, and AB medium. Parallel to this, the Agrobacterium cells are resuspended in the liquid medium containing MS, 30 g/L sucrose, and 20 mg/L AS. This treatment was used as a control in both experiments presented in Fig. 3b. The highest transient GUS expression was observed in 1/10 MS medium containing AS (Fig. 3b), indicating that this treatment significantly increases GUS expression. It was subsequently tested whether the incubation time of Agrobacterium cells in AS-containing 1/10 MS medium affected transient GUS expression. Figure 3c shows that GUS expression increased gradually with increasing incubation time and reached its highest level at 6 h before declining rapidly, suggesting that incubation for 6 h is optimal for transient GUS expression.
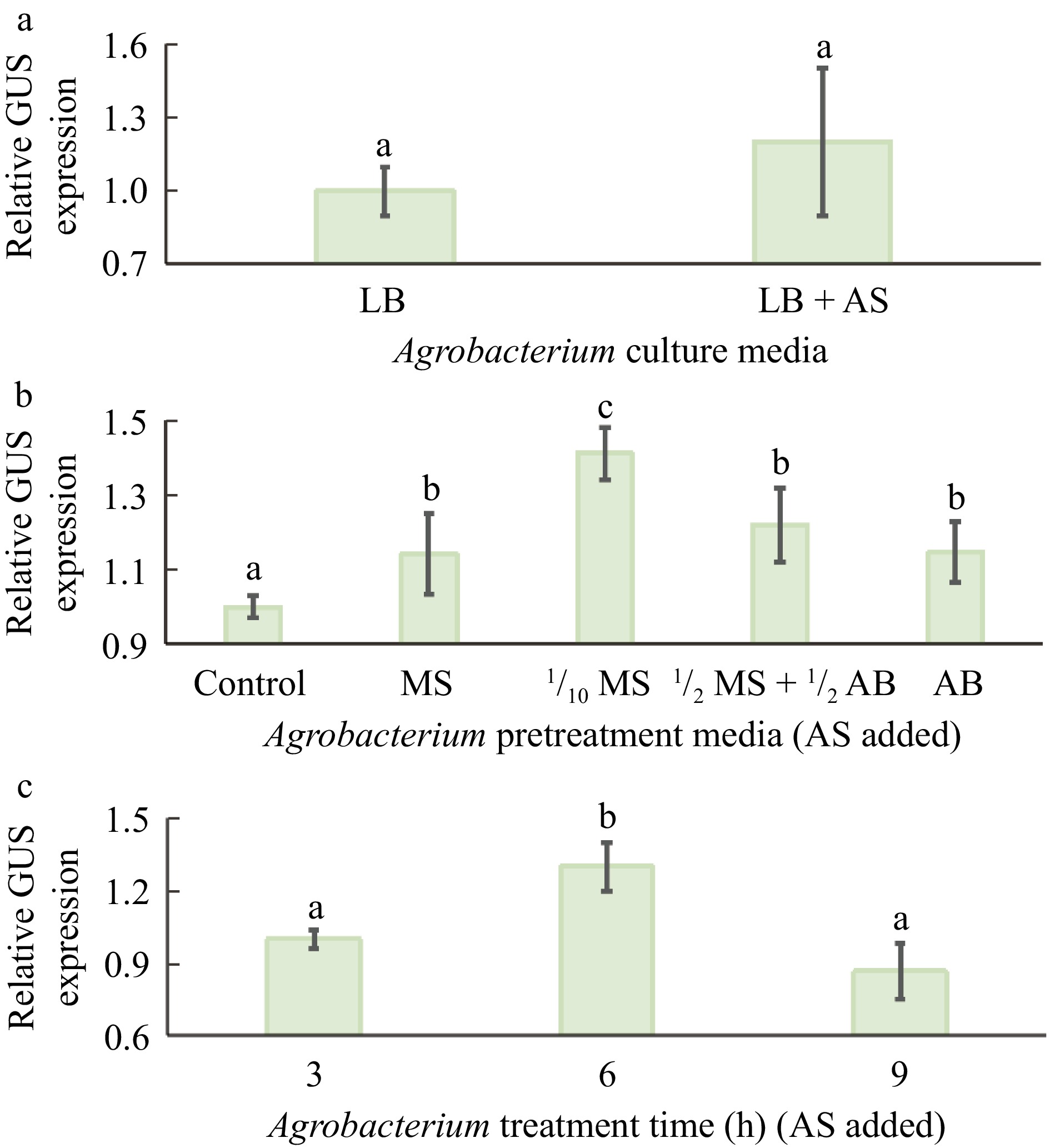
Figure 3.
Effects of components of Agrobacterium cells with acetosyringone (AS) and MS media on Agrobacterium-mediated transient GUS expression. (a) Addition of 20 mg/L AS to the Agrobacterium culture medium, LB medium (pH 7.0). (b) Effects of MS media and Agrobacterium basal (AB) medium on transient GUS gene expression. The duration of the pretreatments of Agrobacterium cells was 3 h. The control was with no pretreatments. (c) Effect of incubation time of Agrobacterium cells in 1/10 MS medium on transient GUS expression. In all experiments, the transient GUS expression activities in the controls were normalized to 1. Different lowercase letters indicate significant differences among different treatments (p < 0.05, ANOVA/LSD).
Effects of sulfamethazine, lipoic acid and paclobutrazol on transient GUS expression
-
Since 3 chemicals, including sulfamethazine (SMZ)[41], lipoic acid (LA)[42,43], and paclobutrazol (PBZ)[37,44] display a promoting effect on stable transformation in various plants, the effects of 3 compounds added to incubation media on transient GUS expression in citrus seedlings were examined. Experiments were conducted using 4-week-old etiolated seedling explants treated with 3 mg/L 6-BA, 1 mg/L 2,4-D, and 0.1 mg/L NAA for 3 h before infection. Agrobacterium cells were resuspended in 1/10 MS medium (pH 5.8) containing 20 mg/L AS and incubated for 6 h before use for infection. The same medium was used for all treatments. Treatment with no added chemicals in the co-cultivation medium was used as a control. It was found that 30 μM SMZ (Fig. 4a), 10 μM LA (Fig. 4b), and 30 μM PBZ (Fig. 4c) were the most effective, resulting in a significant increase in GUS expression, respectively. The combination of any two of these chemicals further increased transient GUS expression, but the combination of all three did not yield a greater increase than any two of the three combined (Fig. 4d). As each chemical functions differently and is inexpensive, a combined use may have synergistic effects on other plant species. We therefore chose to apply all three chemicals to maximize transient expression.
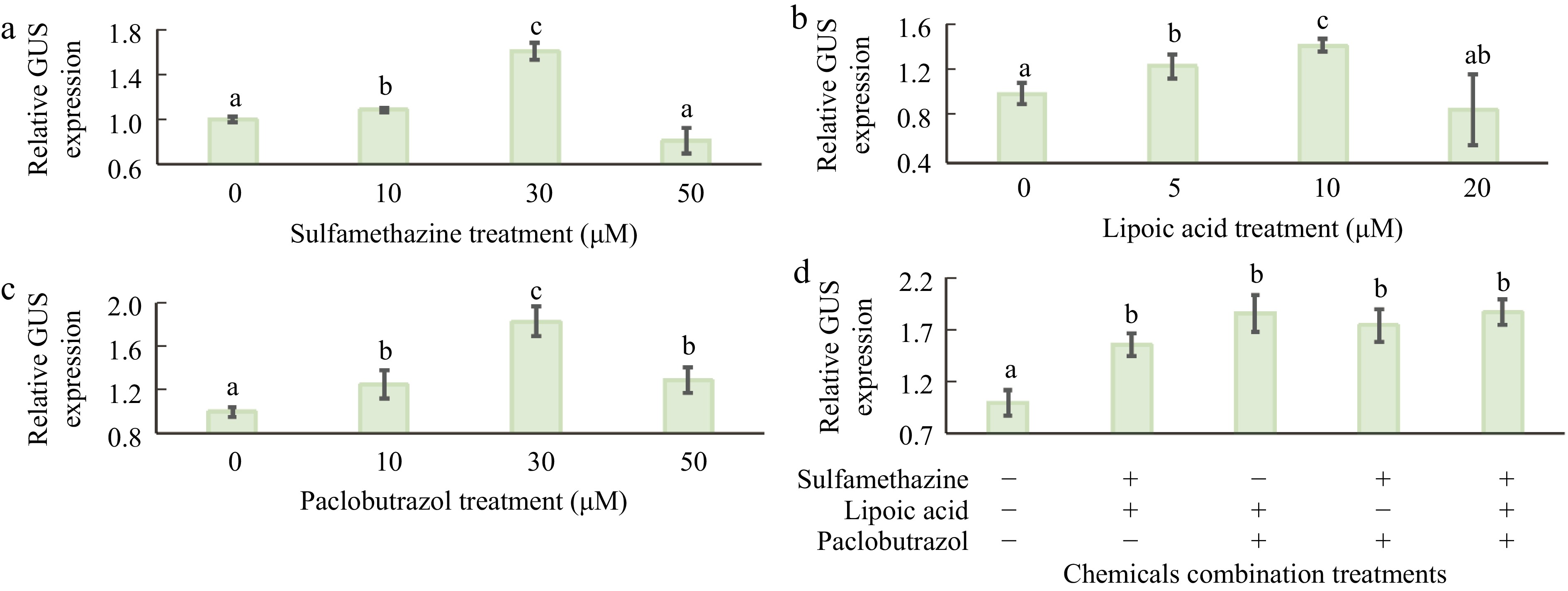
Figure 4.
Effects of chemical treatments added to the co-cultivation media on transient GUS expression. (a) Transient GUS expression in response to sulfamethazine (SMZ). (b) Transient GUS expression in response to lipoic acid (LA). (c) Transient GUS expression in response to paclobutrazol (PBZ). (d) Combined effects of the three chemicals in the co-cultivation medium on transient GUS expression. Experiments were conducted using 4-week-old etiolated seedling explants treated with 3 mg/L 6-BA, 1 mg/L 2,4-D, and 0.1 mg/L NAA for 3 h before infection. Agrobacterium cells were resuspended in 1/10 MS medium (pH 5.8) containing 20 mg/L AS and incubated for 6 h prior to use for infection. Transient GUS expression activity, with no added chemicals in the co-cultivation medium, was normalized to 1. + indicates inclusion of the chemical, while − indicates exclusion. The combination of two of the chemicals (SMZ, LA, and PBZ) can enhance transient GUS expression activity similarly to the effects of all three combined. Different lowercase letters indicate significant differences among different treatments (p < 0.05, ANOVA/LSD).
Synergistic effects of the optimized treatments on transient GUS expression
-
The optimized factors were then combined sequentially and evaluated their synergistic effects on the enhancement of transient GUS expression evaluated. Figure 5 shows that when 4-week-old etiolated explants were combined with the Agrobacterium cell treatment, GUS expression was significantly enhanced. This was further increased by adding the chemical compounds SMZ, LA, and PBZ to the incubation medium, resulting in a five-fold increase in GUS activity in juvenile Carrizo citranges compared to the control, where experiments were conducted using light-grown seedling explants to infect Agrobacterium without any pre-treatments, nor any chemicals in the co-cultivation medium.
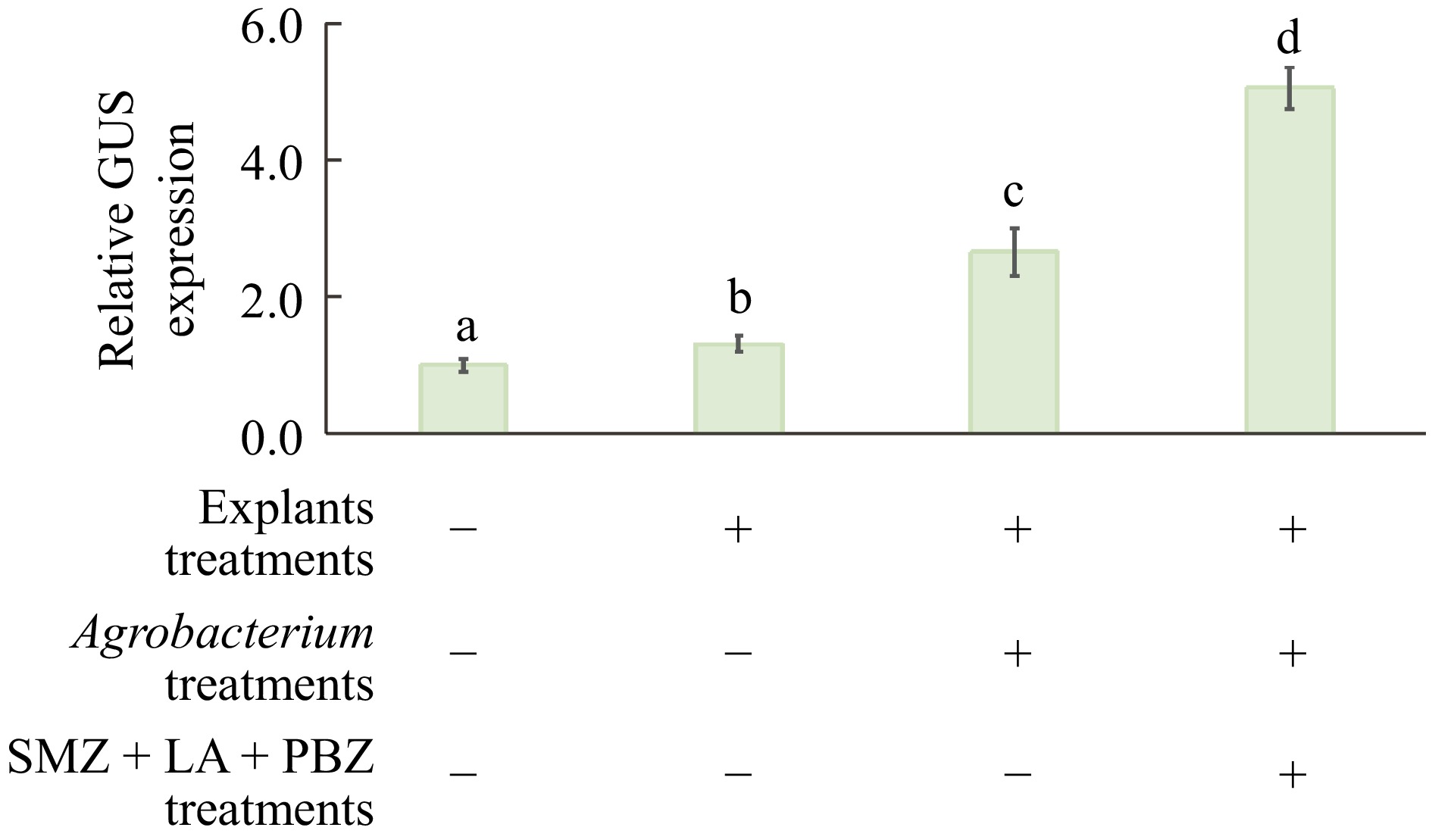
Figure 5.
Combined effects of treatments for Agrobacterium and explants on Agrobacterium-mediated transient GUS expression in citrus epicotyl segments. Expression levels for all other treatments are presented as fold changes compared to the control group. Transient GUS expression activity in the control group (no treatments) was normalized to 1. + or − indicate the inclusion or exclusion of these treatments, respectively. Different lowercase letters indicate significant differences among different treatments (p < 0.05, ANOVA/LSD).
Transient GUS expression enhanced in stem tissues from three mature citrus cultivars
-
To determine whether these treatments, which enhanced transient expression of the GUS expression in juvenile epicotyl tissues could also enhance transient expression in mature shoot tissues, four combinations of previous treatments were tested using mature stem explants of Pineapple sweet orange, Valencia orange, and Washington navel orange. The control refers to the Agrobacterium-mediated mature citrus transformation method described by Orbović et al.[31]. This method utilizes 6-month-old newly developed stem explants from grafted mature buds that are immediately infected with Agrobacterium cells resuspended in an infection medium (pH 5.8) containing MS, 100 mg/L myo-inositol, 10 mg/L thiamine-HCl, 10 mg/L pyridoxine-HCl, 1 mg/L nicotinic acid, 2 mg/L glycine, and 30 g/L sucrose. Transient GUS expression activity in the control was normalized to 1. Figure 6 shows that treatments of Agrobacterium and explants plus any one of the three chemicals increased GUS expression, with a four-fold increase in Pineapple sweet orange (Fig. 6a), a five-fold increase in Valencia orange (Fig. 6b), and a six-fold increase in Washington navel orange (Fig. 6c). However, unlike juvenile epicotyl explants, the combined use of these three chemicals did not result in further increases in GUS expression compared to individual chemical treatments, although each chemical individually enhanced transient GUS gene expression (Fig. 6a−c). Otherwise, similar enhancements were observed in the mature citrus stem tissues.
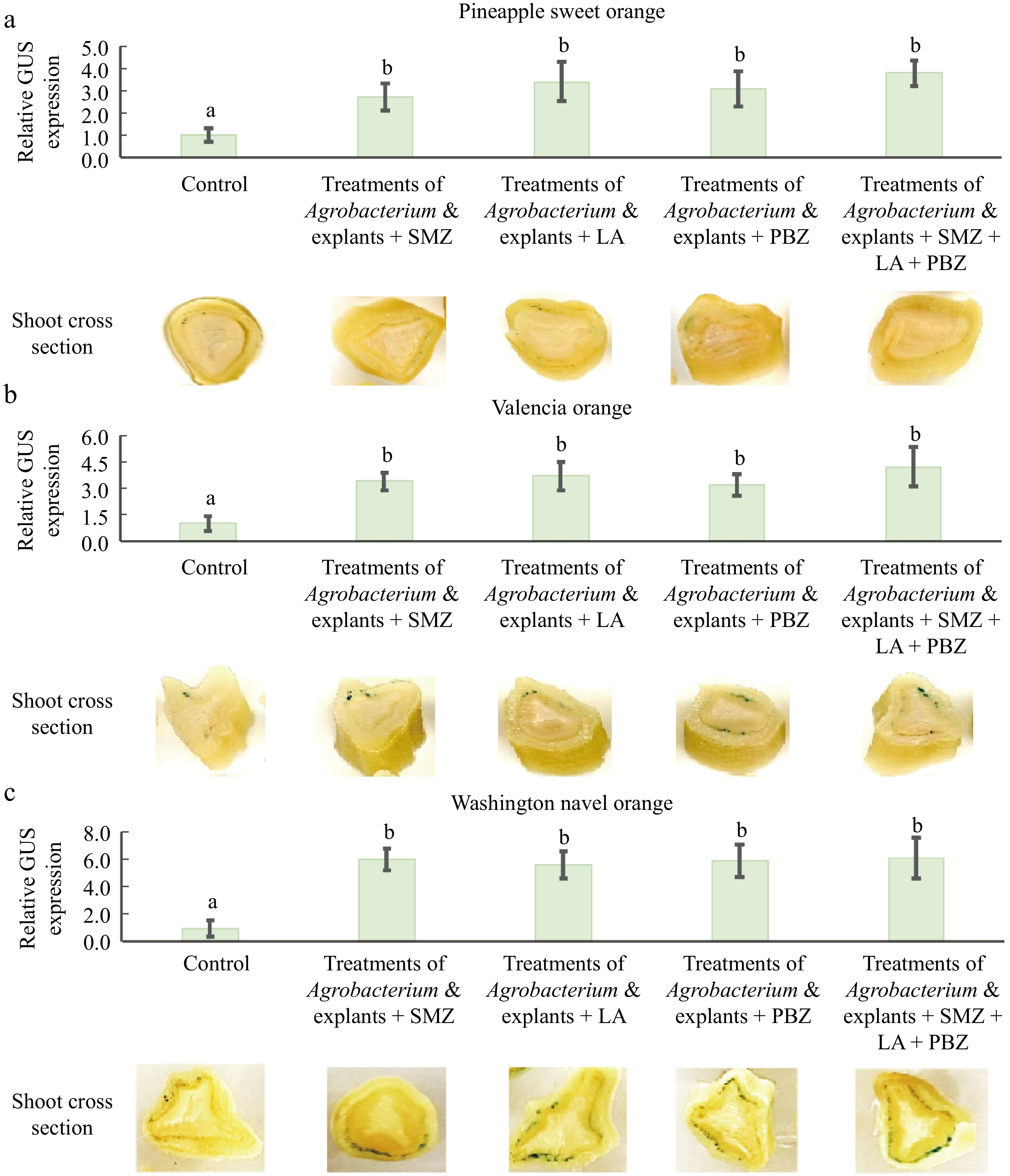
Figure 6.
Combined effects of pretreatments of Agrobacterium and explants on Agrobacterium-mediated transient GUS expression in mature citrus stem tissues. Histochemical staining and quantitative analysis of GUS activities were performed on various treatments in (a) Pineapple sweat orange, (b) Valencia orange, and (c) Washington navel orange. Explant treatment, 6-month-old newly grafted mature stem explants were soaked in a hormone-rich medium containing 3 mg/L 6-BA, 1 mg/L 2,4-D, and 0.1 mg/L NAA for 3 h before infection. Agrobacterium cell treatment, Agrobacterium cells were resuspended in a medium (pH 5.8) containing 1/10 MS and 20 mg/L AS and cultured for additional 6 h before infection. The control refers to the Agrobacterium-mediated mature citrus transformation method described by Orbović et al.[31]. This method utilizes 6-month-old newly developed stem explants from grafted mature buds that are immediately infected with Agrobacterium cells resuspended in an infection medium (pH 5.8) containing MS, 100 mg/L myo-inositol, 10 mg/L thiamine-HCl, 10 mg/L pyridoxine-HCl, 1 mg/L nicotinic acid, 2 mg/L glycine, and 30 g/L sucrose. Transient GUS expression activity in the control was normalized to 1. Different lowecase letters indicate significant differences among different treatments (p < 0.05, ANOVA/LSD).
-
In this study, treatments of Agrobacterium cells and citrus explants before the Agrobacterium and explant co-cultivation, and incorporation of SMZ, LA, and PBZ in the co-cultivation on the Agrobacterium-mediated transient gene expression have been investigated. It has been shown that the combination of these optimized treatments can drastically improve the transient GUS expression in both juvenile epicotyl tissues of Carrizo citrange and mature stem tissues of Pineapple sweet orange, Valencia orange, and Washington navel orange, which will be of significant implication for the improvement of gene editing efficiency in citrus plants. A schematic diagram of these treatments and infection procedure is presented in Fig. 7.
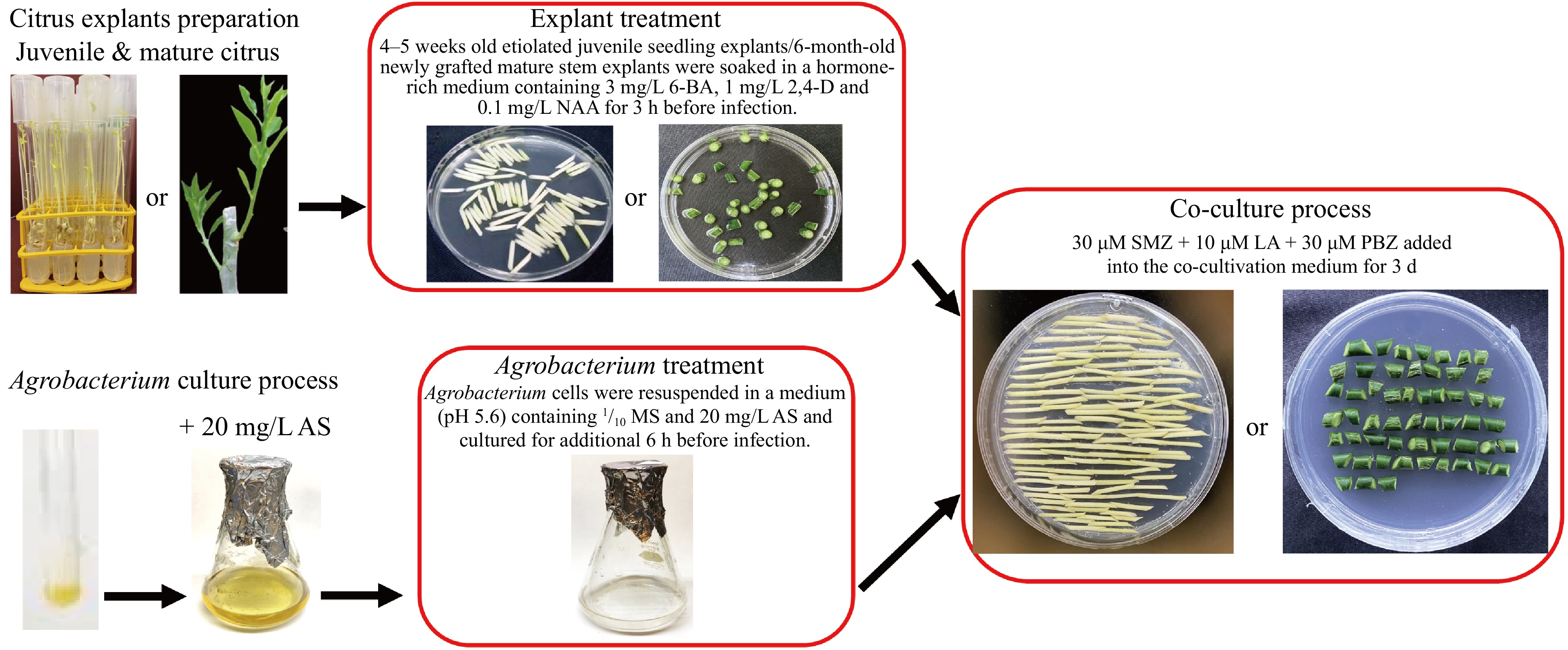
Figure 7.
A summary of the procedure for enhancing Agrobacterium-mediated transient gene expression for both juvenile epicotyl and mature stem tissues of citrus described in this manuscript. The treatments described within the red boxes were incorporated into a conventional procedure for Agrobacterium-mediated infection of both juvenile and mature tissues. The procedure includes treatment of citrus explants and Agrobacterium cells prior to the Agrobacterium-explant co-cultivation, and inclusion of SMZ, LA, and PBZ in co-cultivation medium.
Krenek et al. have demonstrated that developmental stage and the physiological state of plant tissues plays a significant role in T-DNA transfer, integration, and transformation efficiency[1]. The present observations support that these factors also influence the Agrobacterium-mediated transient gene expression activities in citrus tissues. The data show that explants from 4 to 5 week-old seedlings exhibit the highest levels of transient GUS activity compared to other stages of seedlings. Also, the results support that explants from the juvenile seedlings grown in the dark exhibits higher levels of transient gene expression activity when compared to those grown under light conditions, consistent with the results reported by Zhang et al.[45]. Light plays a crucial role in regulating auxin biosynthesis and transport[46,47]. When compared with seedlings grown under light, increased auxin levels in the etiolated seedlings[48] that have been grown in darkness may contribute to higher levels of transient GUS expression observed in the present study.
The efficiency of transgene-free genome editing using Agrobacterium to deliver Cas9 and gRNA largely depends on the transient expression levels of these genes within the T-DNA[25]. Various efforts have been made to improve the stable transformation efficiencies in higher plants[12,49], but relatively little has been done to enhance the transient expression of T-DNA genes. Although several reports have documented the successful production of transgene-free gene-edited citrus plants[50−53], further improvements in the efficiency of transgene-free gene editing in citrus are highly desirable. The present method, which improves the transient expression of T-DNA genes, may therefore be useful for enhancing the efficiency of transgene-free gene editing in citrus and likely in other perennial crops when Agrobacterium cells are used as a delivery vehicle for DNA encoding the Cas9 gene and gRNAs.
The mechanisms by which treating explant tissues with the cytokinin 6-BA and the auxins 2,4-D and NAA before Agrobacterium infection enhances transient gene expression are not clear. It is also interesting to observe that treatment for 6 h or less can increase transient GUS expression, but treatment longer than 6 h does not have the same effect. Similar effects of enhancing transient gene expression by plant hormones have been demonstrated in other plant species previously[54,55].
AS induces expression of vir genes, which are critical for T-DNA transfer into the plant cell nucleus[56,57]. Treating Agrobacterium cells with AS at an appropriate pH and low MS salt concentrations before the plant infection has been shown to increase transient GUS expression[40,56]. T-DNA transfer from the Ti plasmid in Agrobacterium into plant cells is a coordinated action of various vir genes. These vir genes are up-regulated by AS during infection, and AS has therefore been routinely used in plant transformation. In the present study, it was observed that Agrobacterium treated with AS for 6 h before being used for plant infection enhances transient expression of GUS gene in the T-DNA.
One of the significant findings of this study is that all three compounds, SMZ, LA, and PBZ, can enhance transient gene expression in citrus tissues. It has been reported that gene promoters within T-DNA undergo de novo methylation upon Agroinfiltration in leaf tissues[6], which is associated with transcriptional repression or silencing[58] and decreases transgene transient expression[42]. But adding azacitidine (5-AzaC), a methylation inhibitor, to the incubation medium enhanced transient expression[39]. Consistent with earlier findings, adding SMZ, another methylation inhibitor, to the incubation medium also led to an increase in transient GUS expression in both epicotyl and stem tissues (Fig. 5; Fig. 6a−c). This finding again may underscore the importance of demethylation in T-DNAs in promoting transient expression.
Agrobacterium infection often elicits a host defense response, which brings about the generation of excessive harmful reactive oxygen species (ROS)[59], and the addition of antioxidants to incubation mediums often alleviate such response, resulting in the improvement of transient expression efficiency[39]. LA is also an antioxidant and widely used for the enhancement of stable transformation in soybean, tomatoes, wheat, and cotton plants[42,43]. Thus, applying LA to incubation medium likely helps to scavenge excessive ROS accumulated in Agrobacterium-infected plant cells. This mechanism may account for why LA increases transient GUS expression in citrus tissues.
The observation that PBZ increases transient GUS expression in the present case is consistent with that where PBZ promotes stable transformation in Petunia hybrid[44], and citrus[37]. PBZ is an inhibitor of GA biosynthesis[60] but also implicated as a multi-stress ameliorant[61,62]. PBZ may be involved in the mitigation of adverse stress in infected citrus explants and that leads to the improvement of transient gene expression. It was demonstrated that a combination of any two of the three compounds significantly increases transient GUS expression, indicating a positive synergistic interaction.
It is noteworthy that transient GUS expression is highly specific to cambium cells, even under constitutive CaMV35S promoter control. A cambium layer consists of differentiated meristematic cells that produce undifferentiated wood cells inward and phloem/bark cells outward. Due to their active cellular, physiological, and transcriptional activity, it is not difficult to understand why cambium cells express high levels of GUS in epicotyl and stem tissues. The cells derived from the cambium cells, however, become highly differentiated and lose their meristematic characteristics. It may explain why they lose GUS expression activity regardless of how strong or weak the promoter driving the GUS gene is.
The vascular tissue-specific transient expression in citrus tissues might be significant for studies related to CLas, a phloem-colonizing bacteria that has devastated the citrus industry worldwide in recent decades[10,63]. The Agrobacterium mediated-transient expression system reported here along with a nanoparticle-mediated transient silencing of endogenous genes[64] (Li lab unpublished data) could be valuable for the rapid analysis of genes involved in CLas pathogenicity, as well as developmental and growth functions under various conditions, including within graft union regions[65,66].
-
In conclusion, a highly efficient Agrobacterium-mediated transient gene expression method has been developed for both juvenile and mature citrus tissues. This method provides an alternative transient gene expression system for citrus and may also significantly improve the efficiency of Agrobacterium-mediated transgene-free genome editing in citrus and other perennial plant species.
The Citrus Research and Development Foundation (CRDF), USDA Crop Research Initiative Citrus Disease Research and Education (CDRE) and the University of Connecticut. We are grateful to Storrs Agricultural Experiment Station.
-
The authors confirm contribution to the paper as follows: study conception and design: Li Y, Li YJ; data collection: Li YJ, Hu W, Ganie SA, Cheng B; analysis and interpretation of results: Li YJ, Li Y, Liu Z, Duan H; draft manuscript preparation: Li YJ, Li Y, Liu Z. All authors reviewed the results and approved the final version of the manuscript.
-
All data generated or analyzed during this study are included in this published article.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2025 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Li Y, Hu W, Ganie SA, Liu Z, Cheng B, et al. 2025. A novel and efficient Agrobacterium-mediated transient gene expression in citrus epicotyls and mature stem tissues. Fruit Research 5: e002 doi: 10.48130/frures-0024-0035
A novel and efficient Agrobacterium-mediated transient gene expression in citrus epicotyls and mature stem tissues
- Received: 09 July 2024
- Revised: 23 August 2024
- Accepted: 28 August 2024
- Published online: 06 January 2025
Abstract: Agrobacterium-mediated transient gene expression is a powerful technique for rapidly evaluating gene expression in higher plants. This study aimed to improve transient expression levels of T-DNA genes in citrus by investigating the effects of various factors, including seedling age, tissue treatments, Agrobacterium incubation medium and duration, hormone combinations, methylation inhibitors, antioxidants, and more. These parameters were optimized and tested, and significant increases in transient gene expression in juvenile epicotyls of Carrizo citrange and mature stem tissues of Pineapple sweet orange, Valencia orange, and Washington navel orange were observed. The present results demonstrated up to a six-fold increase in transient GUS gene expression, highlighting the effectiveness of these simple and inexpensive treatments. The juvenile and mature citrus explants used in this study displayed high levels of transient expression, which may provide a valuable tool for studying phloem-associated diseases such as Huanglongbing (HLB), facilitating rapid analysis of gene expression involved in Candidatus Liberibacter asiacticus (CLas) pathogenicity. This optimized method may also offer a promising tool for advancing genetic studies and improving the efficiency of Agrobacterium-mediated transgene-free editing in citrus and other economically significant perennial crops.












