-
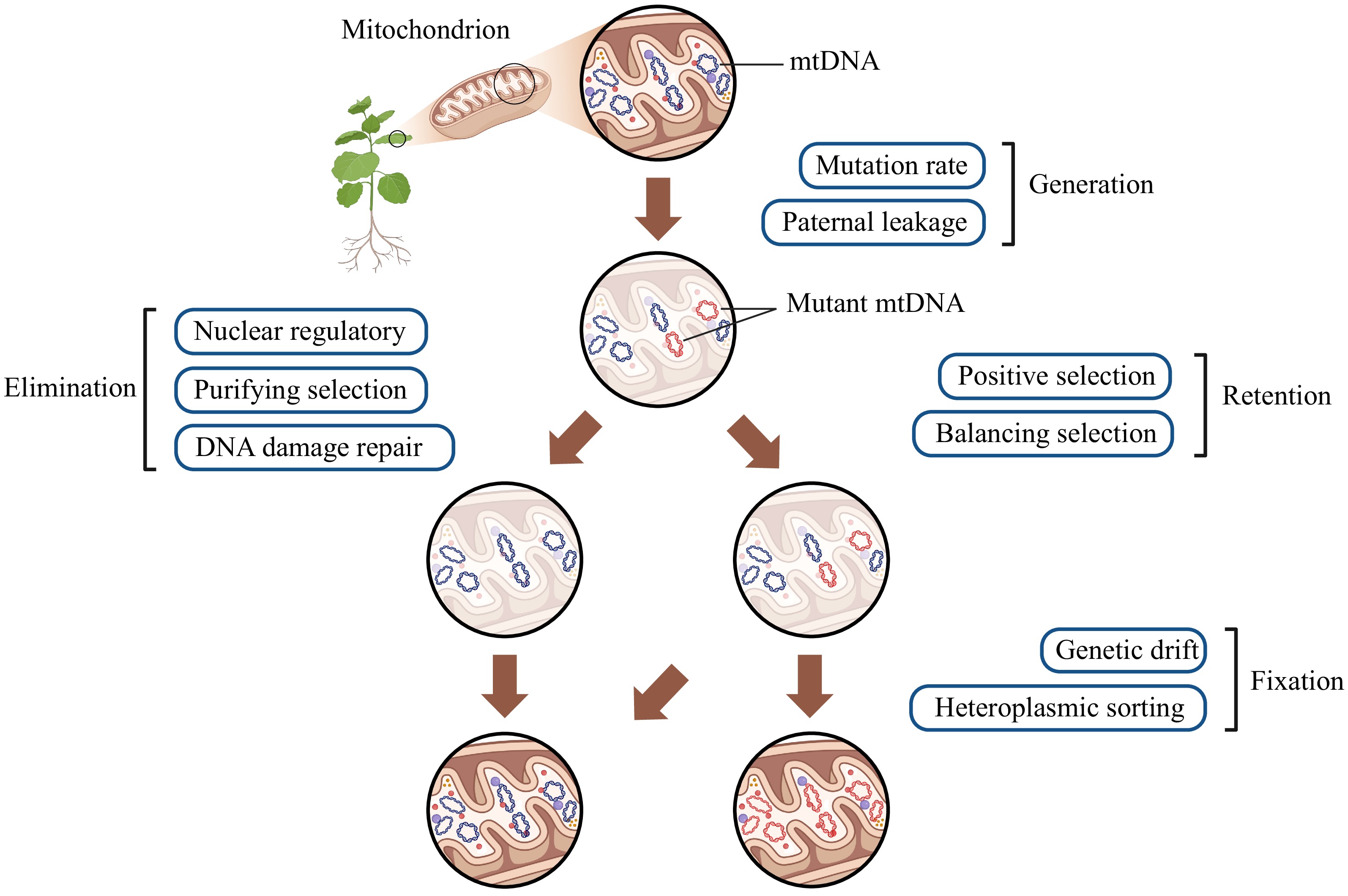
Since Palmer & Herbon[1] noticed the divergences in the mode of mtDNA evolution from six Brassica and Raphanus species, the longstanding mystery that plant mitochondrial DNA (mtDNA) evolves slowly in sequence but rapidly in structure has persisted for nearly 40 years. Subsequent analyses confirmed and extended this paradox. On the one hand, despite encoding similar genes responsible for electron transport chain, plant mtDNA exhibits synonymous nucleotide substitution rates (dS) that are one or two orders of magnitude lower than those in mammalian mtDNA. Additionally, plant mtDNA contains large non-coding regions, in contrast to the smaller and more coding-dense animal mtDNA. Compared to plastid DNA (ptDNA), plant mtDNA exhibits significantly greater structural variability but displays dS less than one-third of those in ptDNA across angiosperms[2,3]. On the other hand, some distant plant taxa independently show surprising accelerations in mtDNA dS, such as Plantago, Silene, Ajuga, and Pelargonium[4−7]. For example, S. noctiflora has experienced a 100-fold increase in dS over the past five million years, and within Plantago, the variation between the fastest and slowest species is about 4000-fold[4,8]. It is largely unknown what mechanism shapes such atypical accelerations, and if any, whether they are shared among these lineages. These observations naturally lead to discussions about how mutations in plant mtDNA sequence and structure are generated, repaired, retained, and fixed. These discussions, in turn, contribute to evolutionary hypotheses to better fit with other genomic characteristics in mtDNA including but not limited to genome size, RNA editing, gene profile, noncoding regions, sparking a broader debate on whether these processes are adaptive or non-adaptive[9−16].
The recent analysis by Zwonitzer et al.[11] published in Proceedings of the National Academy of Sciences USA assessed the connections between organellar genome copy numbers and synonymous nucleotide substitution rates by assuming that fewer genome copies reduce the number of templates available for homologous recombination (HR) that was crucial for repairing mtDNA damage. Their results supported this assumption by showing that mtDNA copy number accounts for 47% of the variation in dS across 60 diverse seed plant species. It is not difficult to understand that low mtDNA copy number will result in a physical limitation for HR because there simply are fewer homologous copies to recombine with. Imagine that there are only 5−10 mtDNA copies but 50−100 mitochondria per cell, each mitochondrion will house << 1 mtDNA copies, and thus effectively less possibility for HR[17]. They also found that mtDNA copy number is negatively correlated with mtDNA size, which could either drive or result from variations in mutation rates. These effects were specific to plant mtDNA and were not observed in ptDNA, although HR repair is also the primary repair pathway in ptDNA (mutations in large inverted repeats (IRs) in ptDNA are less frequent than in single-copy regions). They speculated that the high copy number of ptDNA in leaves might hinder detectable mutations. As for mtDNA, their copy numbers are variable among tissues and developmental stages, with much higher copies in root tips and shoot apex[18], as well as higher copies in mature leaves than in young leaves[19]. Another interesting point is that experimentally removing one IR copy in tabaco (Nicotiana tabacum) can lead to an increase in the total ptDNA copy number. That is, the total amount of ptDNA rather than the number of DNA molecules per organelle regulates ptDNA copy number to ensure a proper gene dosage[20]. Therefore, their analysis suggests that HR influences mtDNA substitution rates in plants and may help to explain mtDNA evolution patterns more broadly across eukaryotes, as this repair pathway is less common in other eukaryotic lineages such as animals.
Zwonitzer's[11] findings are largely in line with the view that plant organellar DNA maintains conserved sequences through a specialized, nearly error-free repair system[1,21]. Specifically, DNA damages such as double-strand breaks (DSBs) can be repaired via various pathways. The HR pathway, which relies on sequences with high similarity to the damaged region (usually other genomic copies or large repeats like the IRs in ptDNA), is the primary repair method in plant mtDNA. In contrast, non-homologous end-joining (NHEJ) and microhomology-mediated end-joining (MMEJ) pathways use limited or no similar sequences, potentially leading to complex genomic rearrangements, insertion, deletions, and duplications[22]. It also inspires interest in interpreting evolutionary differences between coding and non-coding regions of plant mtDNA through differential repair mechanisms, as non-coding regions often show more variations in sequence and structure. However, this interpretation may not be accurate. Christensen[13] once suggested that less accurate mechanisms like break-induced replication (BIR) might predominate in non-coding regions, but subsequent transcript analysis ruled out this possibility[14]. It seems more plausible that the mutation rate is consistent across plant mtDNA. Thus, Christensen[14] further proposed that coding and non-coding regions experience different selection pressures. After being repaired by similar mechanisms (whether through HR or non-HR pathways), coding regions are subject to strong selection that removes most mutations, while non-coding regions experience more relaxed selection, allowing neutral mutations to accumulate. The hypothesis predicts that the nucleotide substitution rate in intergenic regions should be roughly equivalent to the rate of neutral mutations in protein-coding sequences. Research on Fragaria[23] and Arabidopsis thaliana mutation accumulation lines[24] supports this prediction. More analyses are needed, and a great challenge remains in assessing mutation rates in non-coding regions, due to difficulties in aligning whole mtDNA-wide non-coding regions.
Currently, the proteins required for organellar DNA recombination, replication, and repair (RRR) are entirely encoded by nuclear DNA. Therefore, changes in these nuclear genes may affect organellar DNA stability by influencing mutation rates and structural rearrangements. This helps to explain why plant ptDNA and mtDNA share some similar features despite their different origins and functions. For example, if an RRR gene that targets either plastids or mitochondria changes, the corresponding organellar DNA will likely be affected. If an RRR gene bi-targets both organelles, changes in this gene may cause similar alterations in both ptDNA and mtDNA simultaneously. Some studies have observed such patterns[25,26]. However, it is worth noting that experimentally verified functions are still limited to a few genes like msh1[27] and RecA[28], with most predictions based on bioinformatic methods.
Both mtDNA and ptDNA exist in multiple copies within (sub)cells, suggesting that non-adaptive random genetic drift should also be considered. A negative relationship between the size and substitution rate of nuclear DNA and their effective population size (Ne) has been observed across broader eukaryotes, highlighting the impact of genetic drift and selection on nuclear DNA evolution[29]. Zwonitzer et al.[11] observed a similar trend in plant mtDNA, showing a negative relationship between mtDNA copy number and substitution rate, and a positive relationship between mtDNA size and substitution rate. Their findings trigger to turn for the drift-barrier hypothesis arguing that populations with small Ne are less effective at optimizing mutation repair mechanisms compared to those with large Ne, due to low selection efficiency. This inefficiency can result in higher mutation rates and larger genome sizes in species with small Ne, which may explain the accelerated mtDNA dS in some plants[29]. However, Zwonitzer et al.[11] noted that RRR genes responsible for mtDNA stability are constrained by nuclear Ne rather than mtDNA copy number or mtDNA Ne. Therefore, it may be oversimplified to directly use the drift-barrier hypothesis to explain the patterns observed in plant mtDNA. Butenko et al.[15] proposed a non-adaptative drift-burst model to explain gene retention across different eukaryotes. It may help to account for the accelerated mtDNA mutation rates observed in some plants. An apparent burst in mtDNA mutation rates could signal an ancient bottleneck. During such population contractions, mtDNA mutation rate may sharply increase and become quickly fixed, (in a population with small Ne, genetic drift is stronger and selection efficiency is less effective). When mitochondria are temporarily released from strict energy requirements (selection becomes more relaxed), the fixation of mutation can be fixed more rapidly. As a result, the diverse mtDNA mutation rates observed today may stem from these drift-burst events, reflecting the random nature of evolution. Experimental and simulation evidence supports this perspective to some degree. For instance, in msh1 mutants of Arabidopsis, both heterogeneous ptDNA and mtDNA copies can be rapidly sorted when passed on to the offspring. Notably, plastid variants can be lost or fixed even within a single generation that is faster than those in mitochondria[30]. However, a recent study suggested that ptDNA copies can remain in a heteroplasmic state significantly longer than mtDNA copies[31], and it may be one of the possible factors about why plant mtDNA exhibits more sequence variations compared to ptDNA. In addition, organelle bottlenecks are common in various developmental and reproductive processes, which may also contribute to mutation fixation or loss by physically limiting the number of inherited organellar DNA copies[32]. All in all, these observations encourage future research to explore organellar DNA evolution from the perspective of neutral factors.
Moreover, alternations in ways of inheritance may also be one possible reason for accelerated mtDNA in some plants. Although most plant mtDNA is uniparentally inherited, not strict maternal inheritance or paternal leakage can increase the risk of introducing new mutations into offspring mtDNA, and fixing them as new genetic information through positive selection, balancing selection, random drift, etc. These introduced mutations may stimulate the rearrangement of the mtDNA. This perspective uniquely explains the uncoupled evolutionary pattern of ptDNA and mtDNA observed in Silene nutans[33], offering a new perspective on plant mtDNA mutations.
Going back nearly 20 years, Lynch et al.[9] introduced the mutational hazard hypothesis (MHH). According to this hypothesis, genomic differences among mitochondrial lineages are not directly driven by natural selection but by mutation rates. They suggested that very low mutation rates in land plant mtDNA create an environment where noncoding DNA and other genomic changes can accumulate more easily, because excess DNA may pose problems, such as increasing the likelihood of harmful mutations. Consequently, extra DNA tends to accumulate in genomes with low mutation rates, which helps to explain why plant mtDNA has low overall mutation rates but large genome sizes. However, not all studies support the MHH, including those extreme examples with both fast mutation rates and large genomes (e.g., Plantago, Silene, and Pelargonium). Nonetheless, MHH underscores the significant roles of mutation, DNA maintenance, and random genetic drift in shaping organellar DNA.
All in all, no single hypothesis can fully explain the complexity and variability of plant organellar DNA, especially mtDNA mutation, to date. The failure to establish a grand unified theory (GUT) may be frustrating, but it also gives exciting possibilities: the mutations in plant mtDNA likely result from the interaction of multiple factors instead of any single dominant one (Fig. 1). This suggests that we need much more data, particularly from closely related taxa to piece together the puzzle about the mutation of plant organellar DNA.
HTML
-
The authors confirm contribution to the paper as follows: study conception and design: Wu ZQ; analysis and interpretation of results: Wang J, Zou Y; draft manuscript preparation: Wang J, Zou Y, Reeve W, Mower JP, Wu ZQ. All authors reviewed the results and approved the final version of the manuscript.
-
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
This work was funded by the National Natural Science Foundation of China (Grant No. 32170238), the Guangdong Pearl River Talent Program (Grant No. 2021QN02N792), the Chinese Academy of Agricultural Sciences Elite Youth Program (Grant No. 110243160001007).
-
The authors declare that they have no conflict of interest. Dr. Zhiqiang Wu is the Editorial Board member of Genomics Communications who was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of this Editorial Board member and the research groups.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
| Wang J, Zou Y, Mower JP, Reeve W, Wu Z. 2024. Rethinking the mutation hypotheses of plant organellar DNA. Genomics Communications 1: e003 doi: 10.48130/gcomm-0024-0003 |