-
Hepatocellular carcinoma (HCC) stands out as the most prevalent form of primary liver cancer[1]. This type of cancer is particularly significant due to its alarming impact on global health, as it ranks as the fourth leading cause of death attributable to cancer on an international scale. The high incidence and mortality rates associated with HCC underscore the urgent need for enhanced prevention strategies, timely diagnosis, and effective treatment options to combat this formidable disease. The development of HCC is significantly associated with several risk factors, such as infections caused by the hepatitis B virus (HBV), and the hepatitis C virus (HCV)[2,3]. Other contributing elements consist of chronic viral hepatitis, excessive consumption of alcohol, and nonalcoholic steatohepatitis (NASH). Additionally, autoimmune liver diseases, drug-induced liver injuries, and exposure to aflatoxins represent significant risks that can lead to HCC development. These multifaceted risk factors underline the complexity of the disease and the importance of understanding the broader health context in which HCC occurs[4,5]. The prognosis for HCC patients is concerning, particularly in the United States and Asia, where the 5-year survival rates fall between 15% and 38%. This grim statistic is largely attributable to several key factors, including the tendency for late-stage diagnosis, the cancer's resistance to chemotherapy, and the high rates of recurrence and metastasis. One of the main challenges in managing HCC stems from its insidious nature. Many patients experience few to no early symptoms, which leads to significant delays in diagnosis. To mitigate this issue, early screening initiatives targeting high-risk populations are crucial for enhancing the rates of early detection and improving treatment outcomes. Currently, surgical intervention offers a potential curative approach for patients diagnosed with early-stage HCC, particularly for those with a solitary tumor and well-preserved liver function. However, despite the potential for successful surgery, the recurrence rates among these patients can be alarmingly high, reaching up to 70% within 5 years of the operation. Given this challenge, alternative strategies like radiofrequency ablation (RFA) and transarterial chemoembolization (TACE) have surfaced as locoregional therapies intended to decrease the likelihood of surgical recurrence. Unfortunately, many patients are diagnosed with advanced HCC, and it is widely recognized that this stage of the disease shows little sensitivity to conventional chemotherapy[6,7]. Therefore, it is crucial to investigate and create alternative therapeutic strategies for HCC to enhance patient outcomes and increase survival rates. The liver acts as an immune organ, housing a variety of immunocompetent cells. Among these are Kupffer cells (KCs), hepatic sinusoidal endothelial cells (LSECs), hepatic stellate cells (HSCs)[8], pit cells, and lymphocytes, each of which plays a pivotal role in immune regulation and liver pathology. These cells contribute to immune surveillance, inflammation, and fibrosis, and are involved in the progression of liver diseases such as HCC[9,10]. The tumor microenvironment (TME) in HCC consists of cancer cells, various subsets of immune cells, a complex cytokine milieu, and the extracellular matrix, all of which significantly contribute to tumor progression. In view of the unique immunosuppressive microenvironment of HCC, immunotherapy may become a new treatment for patients with advanced HCC. Over the past decade, immunotherapies—particularly those aimed at immune checkpoints, immune checkpoint inhibitors[11,12]. This includes treatments like anti-programmed cell death protein 1 (anti-PD-1), and anti-cytotoxic T-lymphocyte-associated protein 4 (anti-CTLA-4), which have attracted significant attention due to their proven efficacy in treating a wide range of cancers, including melanoma, breast cancer, lung cancer, and liver cancer. Among these, several anti-PD-1 antibodies, such as nivolumab, have received approval from the US[13,14] Food and Drug Administration (FDA) for use in advanced HCC as a second-line treatment, along with pembrolizumab, which is also used in advanced patients. Additionally, a recent clinical trial involving unresectable HCC that evaluated the combination of atezolizumab (anti-PD-L1) and bevacizumab (anti-VEGF) demonstrated improved overall survival (OS), and progression-free survival (PFS) in the combination group compared to those treated with sorafenib alone. However, around 80% of HCC patients do not show an ideal response to immunotherapy[15,16]. Therefore, it is necessary to comprehensively understand the tumor microenvironment and identify the immune cells related to the HCC immunosuppressive microenvironment[17].
MR functions as a strong methodological framework that employs genetic variants as instrumental variables to examine the causal links between different risk factors and the development of diseases[18]. By emulating the parameters of a randomized controlled trial (RCT), MR effectively reduces the influence of confounding variables that are typically encountered in observational studies. This methodological superiority is particularly significant because traditional observational studies aim to identify risk factors by examining the connections between exposures and corresponding outcomes. However, these studies often struggle to establish definitive causal links due to the pervasive issues of confounding and the potential for reverse causality to complicate their findings. In contrast, MR presents a distinct advantage by capitalizing on the fact that genetic variants are assigned randomly at the moment of conception. This random allocation guarantees that these genetic characteristics remain unaffected by any confounding variables that could potentially distort the outcomes in observational research. Furthermore, such a genetic basis makes MR inherently resilient to the problem of reverse causality, thereby strengthening the credibility of the causal inferences drawn from the relationships being studied. Consequently, MR emerges as a powerful tool in the field of epidemiology, offering insights that are less likely to be tainted by the biases that often affect traditional research methodologies[19].
The primary objective of this study was to examine the influence of immune cell characteristics on the progression of HCC by employing a two-sample bidirectional MR analysis. The methodological framework employed by the researchers facilitated a comprehensive investigation into the causal relationships between different characteristics of immune cells and the associated risk of developing HCC. By utilizing this structured approach, the researchers were able to delve deeper into how specific traits of immune cells might influence the likelihood of HCC onset, thereby potentially unveiling critical insights into the mechanisms underlying this type of cancer. This thorough exploration helps in understanding the complex interactions within the immune system and their implications for cancer development, but also to evaluate the potential reverse effect that HCC might exert on those immune cell characteristics. This combined examination offers important perspectives on the complex interactions between immune responses and the progression of cancer, thus deepening our comprehension of the pathophysiological processes involved in HCC[20].
-
This research utilized a two-sample MR method to evaluate the impact of 731 immune cell types on the development of HCC. These immune cell phenotypes were organized into seven distinct categories for comprehensive analysis. MR employs genetic variants as instrumental variables to infer possible causal connections between particular exposures and their related outcomes. To guarantee the strength and credibility of the conclusions drawn from this analysis, it is essential that three key assumptions are fulfilled: first, there must be a strong correlation between the genetic variants and the exposure in question; second, the genetic variants must be independent of any confounding factors; and third, it is essential that the influence of genetic variants on the outcome occurs solely through the specified exposure. For this analysis, data were sourced from GWAS and the FinnGen database. It is essential to highlight that every study incorporated in this analysis obtained the required ethical approvals beforehand to guarantee adherence to ethical research standards. Since the research did not involve the collection of new data from participants, there was no requirement for additional ethical approval, allowing the study to focus solely on the utilization of existing data to derive meaningful conclusions.
Data sources for GWAS of HCC
-
The data regarding HCC was sourced from the FinnGen database, which characterizes HCC as a type of cancer that affects both the liver and the intrahepatic bile ducts. In this comprehensive GWAS, researchers analyzed a substantial cohort of 239,196 individuals of European descent, comprising 518 cases of HCC and 238,678 controls. Through this analysis, a vast number of 155,700,291 SNPs were identified, contributing to the understanding of the genetic factors associated with this malignancy.
Immune cell GWAS data sources
-
The summary statistics for each GWAS associated with immune characteristics can be found in the publicly accessible GWAS directory, particularly for registration numbers GCST0001391 to GCST0002121. This comprehensive study includes a total of 731 immune phenotypes, which are categorized into various types. Among these, absolute counting cells (AC) account for 118 phenotypes. The research additionally examines surface antigens assessed through mean fluorescence intensity (MFI), which includes 389 distinct phenotypes, in conjunction with morphological parameters (MP) that are characterized by 32 phenotypes, as well as relative cell counts (RC) that consist of 192 phenotypes. The phenotypic characteristics related to MFI, AC, and RC include a diverse array of immune cell types. This diversity encompasses B cells as well as circulating dendritic cells (CDCs), which play significant roles in the immune response. These immune cells are critical for the overall functioning of the immune system and contribute to various physiological processes, T-cell maturation, various types of ocytes, bone marrow cells, and TBNK cells, which consist of T cells, B cells, and natural killer cells. Additionally, the regulatory T cell (Treg) panels are also included in this analysis. On the other hand, the morphological parameters (MP) features address the CDC and TBNK panels specifically, providing a detailed lens through which to view immune responses. The initial genome-wide association study concentrating on immune characteristics utilized data gathered from a group of 3,757 participants, mainly of European origin, ensuring that there was no overlap within the study populations. A reference panel created from Sardinian genetic sequences was used to reinforce the findings, analyzing around 22 million SNPs through high-density arrays. Throughout the process, statistical associations were examined while making necessary adjustments for covariates such as sex and age, thereby ensuring the robustness and reliability of the results.
Choice of instrumental variable (IV)
-
We set a threshold of p < 1 × 10−5 for SNPs associated with immune cell phenotypes. These SNPs were then validated through a linkage disequilibrium (LD) check, with parameters of R2 < 0.001 and KB >10000. The values of R2 and KB represent the degree of LD between two loci, indicating that correlated allele frequencies suggest non-independence. For SNPs linked to HCC, we applied a stricter threshold of p < 1 × 10−5 and performed a similar LD check (R2 < 0.001 and KB > 10,000). To assess the strength of the IVs, we calculated the F-statistic for each SNP and excluded those with low F-values (F < 10), which could introduce weak instrument bias. Finally, to minimize the risk of confounding, we used the PhenoScanner database to exclude any SNPs that were associated with potential confounding variables.
Statistical analysis
-
Using the MR-Base platform, we performed GWAS related to immune cells and HCC through an integrated analysis of the data. The analysis of MR was conducted with R software, particularly version 3.4.2, utilizing the 'TwoSampleMR' package. We employed three distinct statistical methodologies to uncover the causal relationship between immune cells and HCC, thereby providing comprehensive insights into how immune traits relate to cancer risk. The methods utilized included IVW analysis, weighted median estimation, and MR-Egger regression. The IVW method seeks to evaluate causal relationships by performing a meta-analysis of the Wald ratios related to each SNP that is part of the research. It is crucial to note that this method operates under the assumption that all SNPs included are valid IVs[21]. On the other hand, MR-Egger regression offers a flexible approach by allowing for the application of the analysis even if some or all of the SNPs do not serve as valid instruments. The slope derived from the MR-Egger regression analysis acts as a crucial marker for indicating the causal link between immune cells and the advancement of HCC. This statistical method, which accounts for potential confounding factors and biases, provides insights into how variations in immune cell populations may influence the emergence and development of this particular type of liver cancer. Thus, interpreting the slope in this context helps to elucidate the role that immune cells may play in the etiology of HCC, reinforcing the significance of investigating immune mechanisms in cancer biology, under the condition that the intercept term is either zero or not statistically significant. Additionally, the weighted median estimator is employed to provide a robust causal estimate, ensuring that at least 50% of the included IVs are confirmed to be valid. This approach involves ranking the effect estimates from each SNP according to their weight, with the median of these weighted estimates being utilized to derive a final causal estimate. A significance threshold of less than 0.05 was set for assessing statistical relevance, thereby assuring the reliability and validity of the results shown[22].
-
In this study, we identified 14 SNPs to serve as IVs, all selected based on a stringent p-value threshold of 1 × 10−5 and an F-statistic > 10 in the GWAS focusing on immune cells. Each of these SNPs showed a connection to HCC at a genome-wide scale. Importantly, of these, eight SNPs displayed a positive relationship with HCC, as depicted in Fig. 1. To obtain more accurate causal estimates, we employed the weighted median estimator, a statistical method designed to ensure that at least 50% of the included instrumental variables are valid. This estimator organizes the effect estimates of each SNP based on their respective weights, and the final causal estimate is computed as the median of these weighted estimates. Through this approach, we discovered a specific immunophenotype, IgD+CD38dimB cells, which was found to have a causal relationship with HCC and posed a significant risk factor for its development. The defining traits of IgD+CD38dimB cells are characterized by their origin from the B cell lineage, along with the expression of IgD and a notably low expression of CD38. On the other hand, our study evaluated an additional 730 immune cells, including CD4+AC, an anti-immune negative regulatory cell that recognizes and selectively targets CD4-expressing T cells, as well as B cell AC. In a similar manner, the study identifies and removes B cells from consideration. Significantly, our research results show that the 730 immune cells that persisted were not linked to the onset of hepatocellular carcinoma (HCC). Furthermore, these immune cells do not appear to contribute to the disease's development or pathogenesis, suggesting their limited relevance in the context of HCC. The OR for IgD+CD38dimB cells and their relationship with HCC was evaluated utilizing the IVW method. This approach provided a strong estimation of the causal link between these specific immune cell characteristics and the risk of developing HCC. The resulting OR was identified as 1.346, with a 95% CI ranging from 1.079 to 1.680, and a p-value of 0.008. This finding suggests a statistically significant correlation between IgD+CD38dim B cells and HCC incidence. In addition to the IVW method, several other MR techniques were employed to derive causal estimates regarding the association between immune cell traits and HCC. Among these methods, the weighted mode method indicated an OR of 1.355, accompanied by a 95% CI of 0.999 to 1.838 and a p-value of 0.072. Although this result hints at a possible relationship between immune cells and HCC, it did not achieve statistical significance. Similarly, the weighted median method yielded an OR of 1.344 with a 95% CI from 0.972 to 1.858 and a p-value of 0.072, reaffirming the trend observed in the weighted mode method, yet also falling short of reaching statistical significance. The MR-Egger regression analysis resulted in an OR of 1.238, which was paired with a 95% CI that spanned from 0.896 to 1.710. The p-value associated with this analysis was reported as 0.219. Although these findings may indicate a possible association between the variables under study, it is crucial to emphasize that the p-value does not reach a threshold of statistical significance, thus suggesting that the evidence is not robust enough to draw firm conclusions. In a similar vein, the simple mode method also provided an odds ratio of 1.303, which came with a 95% confidence interval ranging from 0.770 to 2.203, and a p-value of 0.341. This further corroborates the notion that while there is an observed effect, the level of statistical significance remains insufficient to validate a definitive connection. This also supports the notion of a potential association, yet it too falls short of reaching statistical significance. Detailed results from these analyses are illustrated in Table 1 and Fig. 2. Despite the p-value for the MR-Egger analysis exceeding the threshold of 0.05, all four analytical methods employed in this study produced positive beta values. This consistency across the various methodologies points toward a shared trend of a positive association. Such a consistent pattern signals that the analyses yield similar directional conclusions regarding the relationship being examined. A fundamental distinction between the MR-Egger method and the IVW approach lies in the inclusion of an intercept term in the MR-Egger regression. This intercept serves to account for potential pleiotropic effects among the IVs. Specifically, the intercept in the MR-Egger regression gauges the average effect of polymorphisms, while the slope offers an unbiased estimate of the causal relationship in question. Typically, the intercept assists in detecting the presence of pleiotropy, whereas the slope accurately reflects the underlying causal link between exposure and outcome. The IVW method, in contrast, demonstrates a smaller standard error than that of the MR-Egger method, indicating a higher degree of precision in the causal estimates produced. When heterogeneity and horizontal pleiotropy are absent, the results arising from the IVW method are deemed the most trustworthy and are considered the gold standard in MR analyses, largely due to their precision and efficiency. This analysis produced statistically significant results, which are further detailed in Table 1 and Fig. 2.
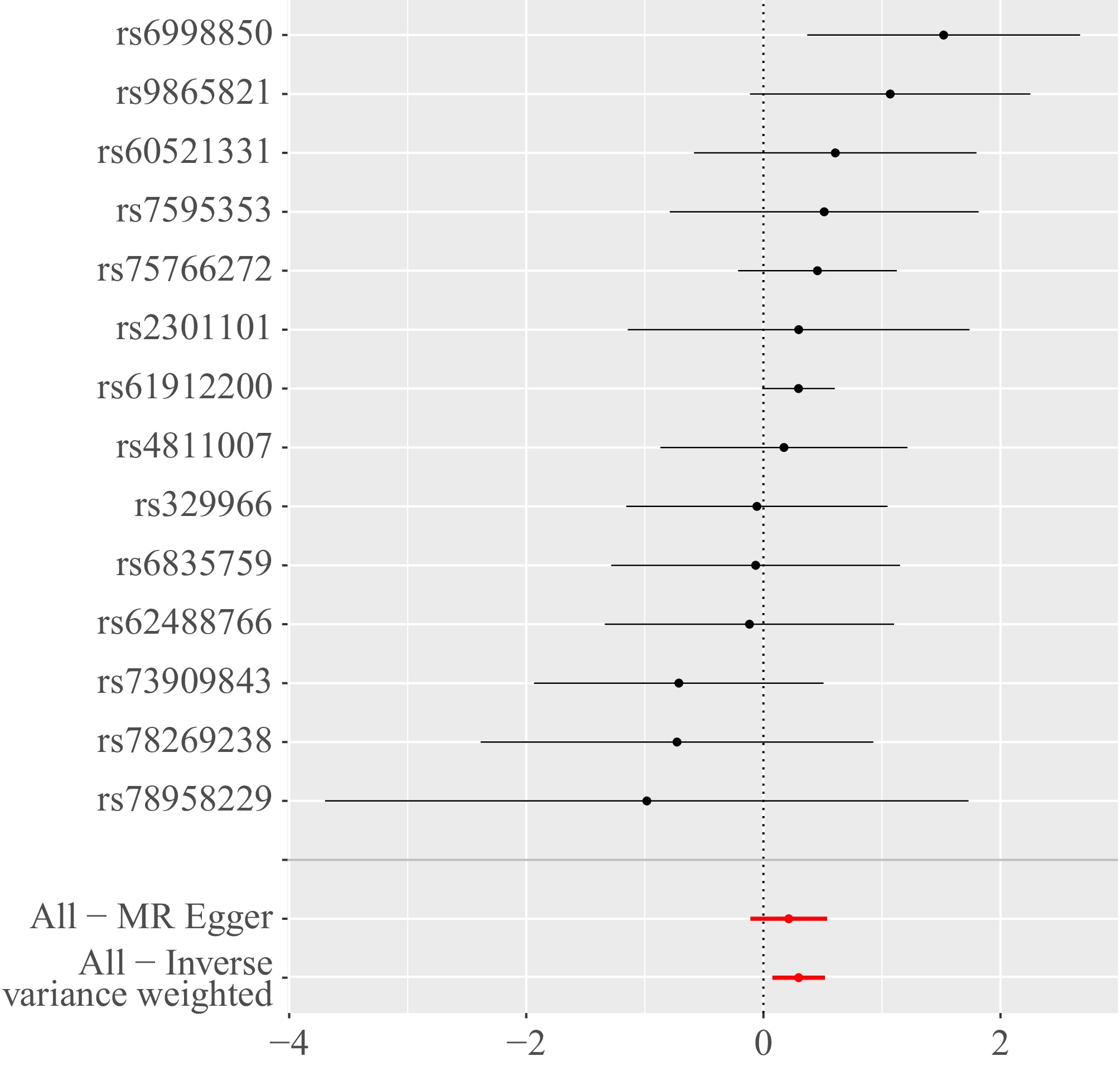
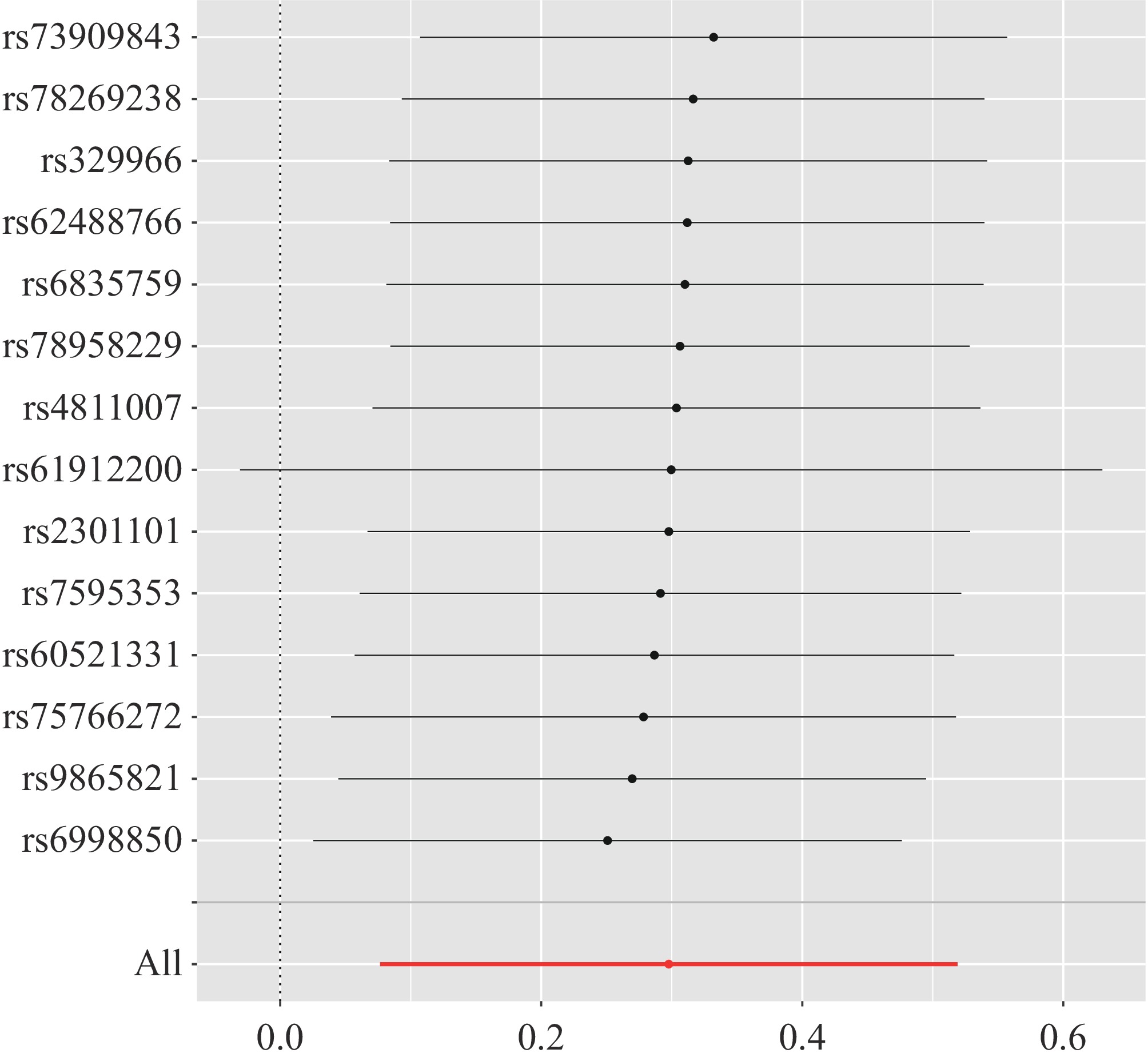
Figure 1.
Forest plot of the causal effects of single nucleotide polymorphisms associated with immune cells on hepatocellular carcinoma. The significance of the red lines are MR results of MR-Egger test and the IVW method.
Table 1. MR estimates from each method of assessing the causal effect of immune cells on the risk of hepatocellular carcinoma.
MR method Number
of SNPsBeta SE p-value OR 95% confidence interval I2 Cochran Q statistic Heterogeneity p-value Horizontal
pleiotropy p-valueMR-Egger 14 0.213 0.164 0.219 1.238 0.896−1.712 0.908 12.254 0.425 0.036 Inverse variance weighted 14 0.297 0.112 0.008 1.346 1.079−1.680 0.556 12.761 0.466 0.039 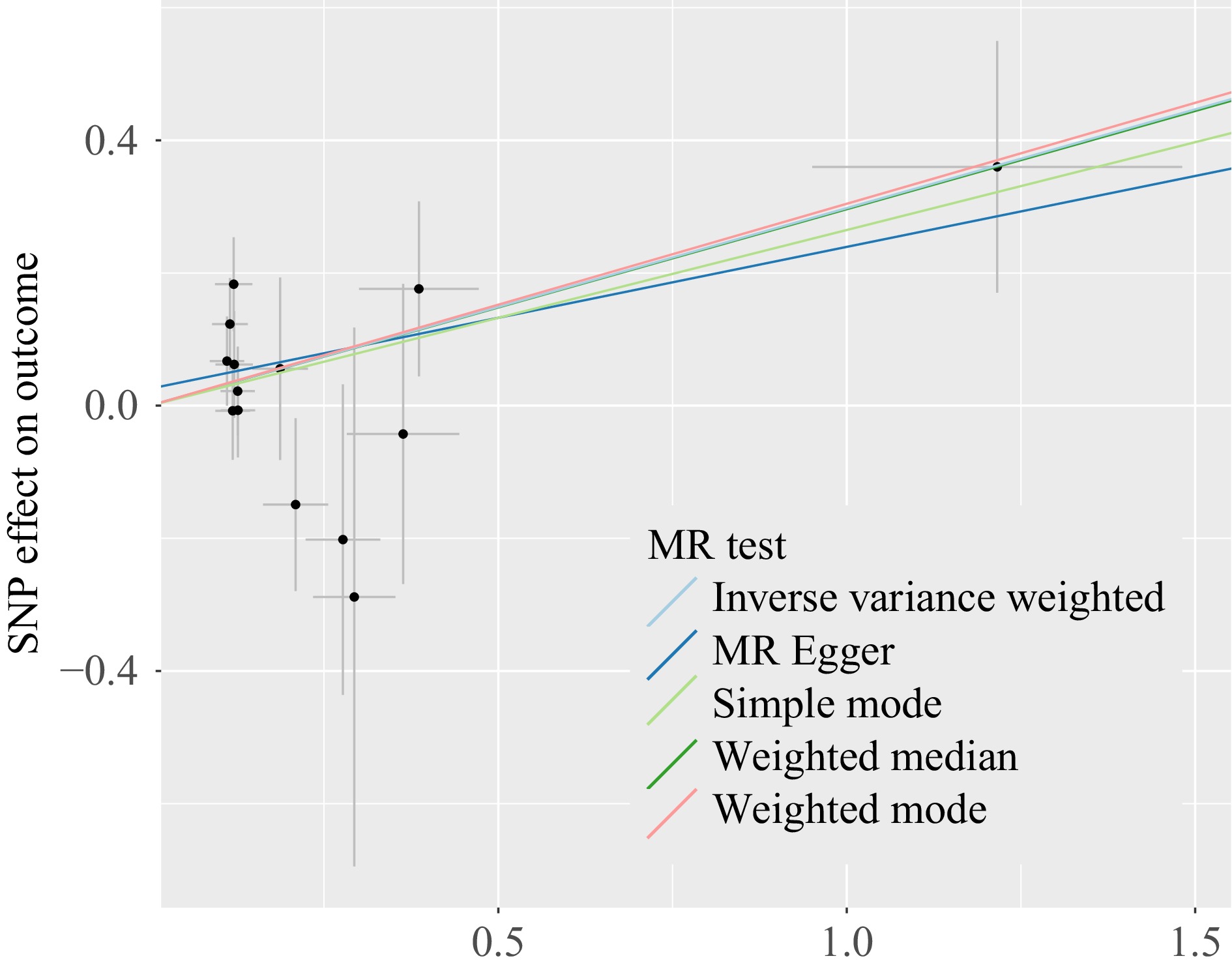
Figure 2.
Scatter plots of genetic associations with immune cells against the genetic associations with hepatocellular carcinoma. The slopes of each line represent the causal association for each method. The Cambridge blue line represents the inverse - variance weighted estimate, the dark green line represents the weighted median estimate, the dark blue line represents the Mendelian randomization - Egger estimate, the pink line represents the Mendelian randomization - weighted mode, the light green line represents the Mendelian randomization - simple mode.
Sensitivity analysis
-
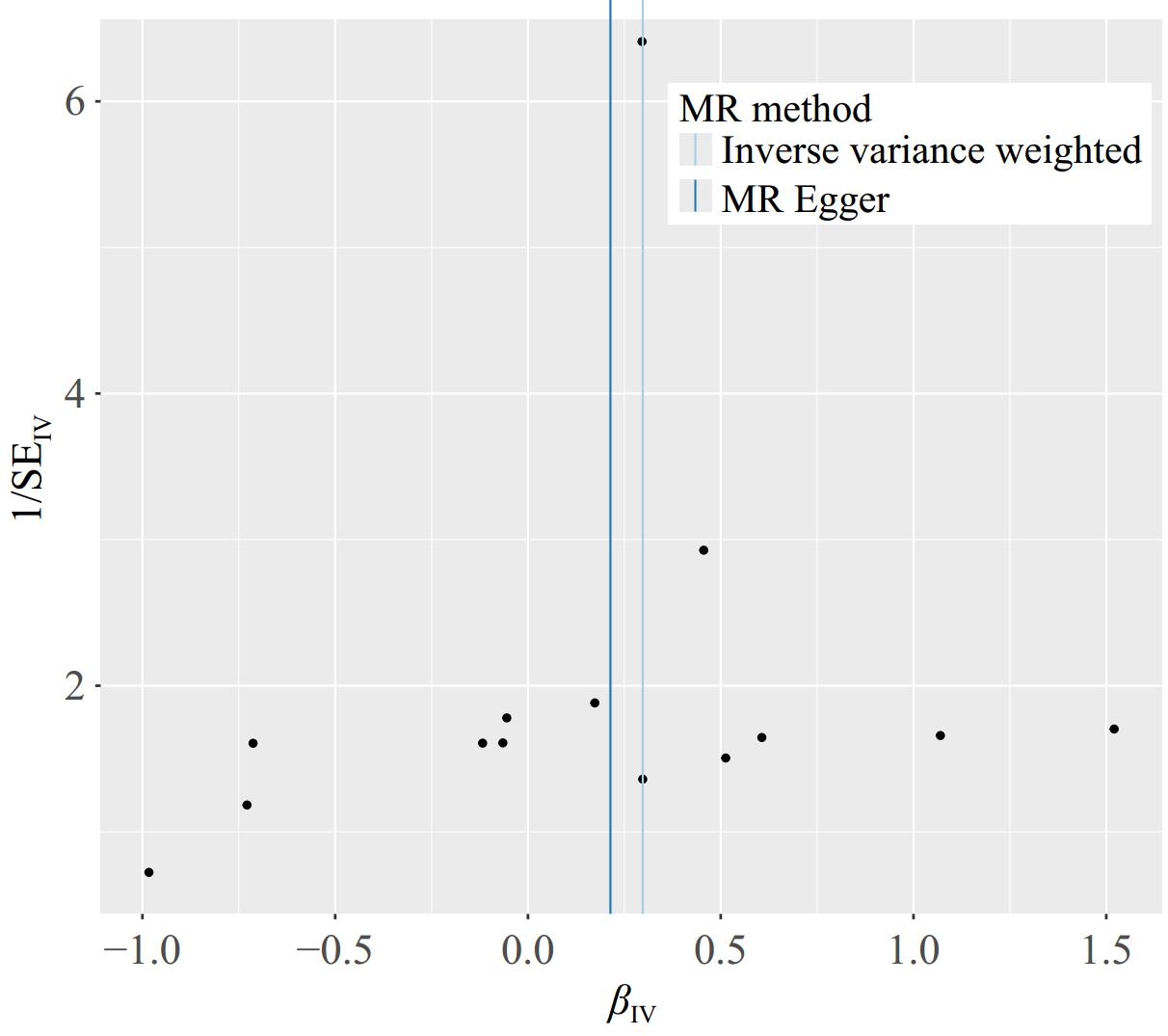
Initially, the heterogeneity function was employed to assess the presence of heterogeneity among immune cells in relation to HCC. The findings indicated an absence of heterogeneity in this context, as highlighted in Table 1. Additionally, a funnel plot was utilized to evaluate the heterogeneity of SNPs. The plot revealed that the distribution of points on either side of the IVW line displayed a roughly symmetrical pattern, with no notable outliers detected, as illustrated in Fig. 3. Furthermore, an investigation into horizontal pleiotropy was conducted using the MR-PRESSO method. The outcomes of this analysis confirmed that there was no evidence of horizontal pleiotropy between immune cells and HCC, further corroborated by the data presented in Table 1. To guarantee the reliability and strength of the results obtained, a sensitivity analysis was conducted using the leave-one-out methodology. This approach involves systematically omitting one observation from the dataset at a time to assess how much influence each individual data point has on the overall outcome. By implementing this method, we can gain valuable insights into the stability of our findings, as well as identify any potential outliers or influential points that may disproportionately impact the results. The application of this technique is crucial for validating the conclusions drawn from the data and ensuring that they are not unduly affected by any singular observation. This approach demonstrated that the overall error line exhibited minimal variation upon the exclusion of each SNP. All error lines remained positioned to the right of zero, as shown in Fig. 4. In summary, the findings derived from the MR analysis in this study are deemed reliable. The consistency of results across various MR methods, along with the lack of significant biases attributed to heterogeneity or pleiotropy, supports the interpretation that there may be a causal relationship between the characteristics of immune cells and the progression of HCC.
-
Gu et al.[22] discovered a distinct population of IgD+/CD38+ small B cells present in cases of necrotizing lymphadenitis and systemic lupus erythematosus, which implies that these anergic or autoreactive B cells might play a detrimental role in disease pathology[23]. This finding raises important considerations about the functional implications of these B cell subsets in autoimmune conditions and how their presence could contribute to disease progression[24]. Research led by Garnelo et al. highlighted that CD38 is expressed not only on activated B cells, which include germinal center B cells, memory B cells, plasmablasts, and plasma cells, but is also found on various other immune cells such as T cells, natural killer (NK) cells, and monocytes following their activation[21]. The expression of CD38 on these immune cells underscores its significance in understanding immune responses and its potential involvement in tumor immunology, particularly in HCC. Notably, the density of CD38+ tumor-infiltrating lymphocytes (TILs) was found to correlate with the survival rates of patients suffering from HCC, suggesting that CD38+ TILs may have implications for patient prognosis. Research conducted by the Zhongshan Hospital affiliated with Fudan University, led by Academician Jia Fan, indicated that in the context of liver cancer, the immune microenvironment and immune therapy strategies predominantly feature B cells characterized by an IgD-IgG-CD27-CD38-memory phenotype[25]. These memory B cells are involved in orchestrating anti-cancer responses through the release of interferon-gamma and granzyme B, thus interacting cooperatively with CD8+ T effector cells to enhance the anti-tumor immune response[26]. This cooperative interaction is vital for developing therapeutic strategies against liver cancer. Using a Bayesian-weighted MR approach, Zhu et al. found that B-cell activator receptors on IgD+ CD24-B cells appear to reduce the risk of developing HCC, with phenylacetylglutamate acting as a mediating factor. This discovery reinforces the intricate role that specific B cell subsets play in modulating immune responses and indicates possible protective mechanisms against cancer development[27]. Moreover, previous investigations have indicated that CD38 is present in specific tumor subsets, particularly those exhibiting elevated levels of immune cell infiltration, either due to basal activity or as a result of treatment. Tumors that have been subjected to PD-1/PD-L1-targeted therapies may develop treatment resistance, potentially facilitated by the tumor microenvironment through the release of all-trans retinoic acid and interferon-beta (IFN-β), which can lead to the up-regulation of CD38. Interestingly, earlier studies did not find a direct association between IgD+CD38dim B cells and the development of HCC, suggesting a more complex network of interactions that needs further exploration. The primary aim of this study was to investigate the influence of immune cells on the development of HCC through a two-sample MR analysis utilizing data from GWAS. The findings indicated a significant association between B cells co-expressing IgD and CD38 and an increased risk of HCC, highlighting their potential involvement in the pathogenesis of this liver cancer and suggesting avenues for further research into their role in tumor-related immune responses.
This study does, however, present certain limitations that must be acknowledged. Firstly, the precise mechanism underlying the causal relationship between IgD+CD38dimB cells and liver cancer remains unclear and necessitates further exploration. A comprehensive understanding of this pathway is essential for elucidating how these immune cells contribute to the development of liver cancer, and subsequent research efforts should aim to clarify this connection. Secondly, the process of verifying genetic polymorphisms poses significant challenges, and there remains a possibility of misclassification regarding these variants. Even with the application of the MR-Egger method aimed at addressing potential biases, the risk of inaccuracies cannot be entirely ruled out[27]. Hence, careful consideration must be given to the methods used for genetic analysis to ensure reliable results. Furthermore, the immune cell dataset extracted from the GWAS in this study was based on a mixed population, while the liver cancer data was specifically obtained from a European cohort. This discrepancy between the populations could introduce potential bias due to population stratification, ultimately leading to uncertainty about the generalizability of the findings across different ethnic or demographic groups[28]. Consequently, these results signal the need for additional research to confirm their applicability beyond the studied populations. Lastly, the issue of overidentification in two-sample MR studies may arise, which could result in an exaggerated estimation of the association between SNPs and exposure levels. This concern highlights the importance of employing meticulous methodologies in future research to avoid any misleading conclusions regarding correlation and causation in the context of genetic and health associations.
-
The findings of the current study provide strong evidence derived from MR analysis, suggesting that IgD+CD38dim B cells may have a causal relationship with an increased risk of HCC. This indicates that these specific immune cells could serve as an important risk factor in the development of hepatocarcinogenesis, highlighting their potential role in the pathophysiology of this type of cancer.
-
All relevant studies were approved by the local ethics committee. Since this study did not involve the collection of new data, it was not necessary to apply for ethical approval.
-
The authors confirm contribution to the paper as follows: conceptualization, methodology, software, validation: Chen Y, Chen H; writing - original draft: Chen Y, Chen H, Lin J; writing - review & editing: Deng Y, Lin J; supervision: Deng Y, Lin J, Lin R; resources: Lin J, Peng L, Gong G; investigation: Lin J, Peng L; data curation: Lin J, Peng L, Xiao S; formal analysis: Xiao S; project administration: Lin R; visualization: Gong G. All authors reviewed the results and approved the final version of the manuscript.
-
The data of this study were obtained from the public GWAS database and FinnGen database.
-
We thank Mu-lan Cai for statistical support of this article.
-
The authors declare that they have no conflict of interest.
-
# Authors contributed equally: Yun Chen, Hongbin Chen, Jinrong Lin
- Copyright: © 2025 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Chen Y, Chen H, Deng Y, Lin J, Peng L, et al. 2025. Effect of immune cells on hepatocellular carcinoma: a two-way Mendelian randomization study. Gastrointestinal Tumors 12: e006 doi: 10.48130/git-0025-0006
Effect of immune cells on hepatocellular carcinoma: a two-way Mendelian randomization study
- Received: 06 September 2024
- Revised: 22 January 2025
- Accepted: 13 February 2025
- Published online: 31 March 2025
Abstract: The objective of the present study was to investigate the effect of immune cells on hepatocellular carcinoma (HCC). The GWAS Catalog database and FinnGen database were used to obtain extensive genomic data on immune cells and liver cancer. Single nucleotide polymorphisms (SNPs) associated with immune cells as instrumental variables (IV). A two-sample MR study method was used to detect the effect of immune cells on hepatocellular carcinoma. The results were analyzed using five statistical methods: inverse variance weighting (IVW), weighted median estimation, weighted model, simple model, and MR-Egger regression. In the GWASs of immune cells, 14 SNPs were selected as the instrument variables under the conditions of p < 1 × 10−5, R2 < 0.001, physical distance >10,000, and F > 10. It was found that Bcells with both IgD+ and weak CD38 expression were risk factors for HCC development. The odds ratio (OR) of IgD+CD38dimB cells in HCC calculated by the IVW method was 1.346 (95% CI: 1.079−1.680; p = 0.008). The results of other MR methods were as follows: Weighted mode (OR = 1.355; 95% CI: 0.999−1.838; p = 0.072), weighted median (OR = 1.344; 95% CI: 0.972−1.858; p = 0.072). Sensitivity analysis showed that there was no heterogeneity and horizontal pleiotropy in the results. The MR analysis method showed that IgD+CD38dimB cells are risk factors for HCC.
-
Key words:
- Hepatocellular carcinoma /
- Immune cells /
- Mendelian randomization