HTML
-
Diatrypaceae consists of 22 genera and approximately 1035 species (Wijayawardene et al. 2020, Hyde et al. 2020b, Konta et al. 2020; Dissanayake et al. 2021). The taxa are generally characterized by perithecial ascomata immersed in a stroma, asci with long pedicels, allantoid ascospores and coelomycetous or hyphomycetous asexual morph (Shang et al. 2017, Hyde et al. 2020b). The asexual morphs of Diatrypaceae are morphologically similar and thus cannot be used for taxonomic differentiation (Acero et al. 2004). Genera in Diatrypaceae are delineated mainly based on stromatal morphology e.g., pustulate, valsoid, eutypoid, well-developed or poorly-developed (Vasilyeva & Stephenson 2005, Senwanna et al. 2017). Early treatments of Diatrypaceae were based on morphology, while recent studies have included phylogenetic data (Mehrabi et al. 2016, Dayarathne et al. 2020, Konta et al. 2020).
Diatrypella was introduced as a segregate of Diatrype with polysporous asci (Croxall 1950). Characteristics of Diatrypella include conical or truncate, discoid or cushion-like stromata delimited by a black zone on the host tissues, perithecial ascomata, umbilicate or sulcate ostioles, and numerous ovoid to allantoid ascospores. The asexual morph is described as libertella-like (Senwanna et al. 2017, Shang et al. 2017, Hyde et al. 2020a). Diatrypella includes approximately 115 species (Wijayawardene et al. 2020). In the last decade, several new species have been introduced using evidence from morphology coupled with ITS and β-tubulin sequence analyses. These include D. iranensis, D. macrospora, D. tectonae and D. yunnanensis (Mehrabi et al. 2015, 2016, Shang et al. 2017, Hyde et al. 2020a). Molecular studies, have however, revealed that Diatrypella is polyphyletic (Senwanna et al. 2017). Acero et al. (2004) suggested that polysporous asci is a characteristic that evolved independently multiple times during evolution of Diatrypaceae (Acero et al. 2004, Senwanna et al. 2017). This suggests that current features used in the taxonomy of Diatrypaceae may not reflect the evolutionary history (Mehrabi et al. 2016), hence highlighting the importance of using molecular data for delimitation of genera within the family.
Diatrypella taxa have broad geographic distribution as saprobes (e.g. D. heveae, D. quercina, D. yunnanensis), endophytes (e.g. D. favacea, D. frostii) or occasionally as suspected pathogens (e.g. D. japonica, D. vulgaris) mainly on woody angiosperms (Pitt et al. 2013, de Almeida et al. 2016, Senwanna et al. 2017, Rudolph et al. 2018, Rashmi et al. 2019, Hyde et al. 2020a, Li et al. 2020). Some species have a broad host range, others such as D. vitis (on grapevines) have been reported only on one host genus (Acero et al. 2004, Farr & Rossman 2020).
In the present study we report a new host and geographical record of Diatrypella macrospora on Quercus cerris from Italy based on combined ITS and β-tubulin sequence analysis. Our comprehensive analysis also highlights the need for extensive revisions within Diatrypaceae.
-
Sample collection, morphological studies and isolation
A dead land branch of Quercus cerris was collected in the Province of Forlì-Cesena, Italy in July 2019. Samples were brought to the laboratory and kept in paper envelopes. Macroscopic characters (enlarged host surface and ascomata) were examined using a Motic SMZ 168 series stereomicroscope. Microscopic features were observed using a Nikon DS-Ri2 digital camera fitted to a Nikon Eclipse 80i compound microscope. Thin cross sections of stromata were prepared manually and mounted in water on glass microscopic slides. Photomicrographs were prepared with Adobe Photoshop CC v. 20.0.5 (Adobe Systems, USA) and all character measurements were made with Tarosoft Image FrameWork v. 0.9.0.7.
Single ascospore isolation was carried out as described in Senanayake et al. (2020). After germination, ascospores were transferred aseptically to malt extract agar (MEA) medium and incubated at room temperature. A pure culture was obtained and its characteristics were also observed. Dried specimens and living cultures were deposited in the Mae Fah Luang University herbarium, Thailand (MFLU) and the Mae Fah Luang University Culture Collection (MFLUCC) respectively.
DNA extraction, PCR amplification and phylogenetic analyses
The methods used in DNA extraction, PCR amplification, sequence analyses and genetic analyses are as outlined in Dissanayake et al. (2020) with the following or modifications. The internal transcribed spacer (ITS) and β-tubulin loci were amplified by polymerase chain reaction. The primers used for amplification were: ITS5 and ITS4 (White et al. 1990) for ITS and T1 and Bt2b (Glass & Donaldson 1995, O 'Donnell & Cigelnik 1997) for β-tubulin. The PCR conditions for both ITS and β-tubulin were set as follows: initial denaturation of 94℃ for 3 mins, followed by 35 cycles of denaturation at 94℃ for 30s, annealing at 55℃ for 50s, elongation at 72℃ for 1 min and final extension at 72℃ for 10 mins. DNA sequencing was performed at BGI Shenzhen, China. The closest related strains and sequences spanning the diversity of Diatrypaceae were compiled following recent publications (Dayarathne et al. 2020, Konta et al. 2020) (Table 1). For the Maximum likelihood (ML) analysis, the optimal ML tree was obtained using 1000 separate runs and GTR+GAMMA was used as the model for nucleotide substitution. For Bayesian inference analyses (BI), two parallel runs, each consisting of four simultaneous Markov chains were executed for 4, 000, 000 generations, sampling one tree every 1000th generation. 25% of the trees were discarded as the burn-in phase in the analysis. The remaining trees were used to calculate posterior probabilities in the majority rule consensus tree. Convergence was determined when the average standard deviation of split frequencies reached 0.01.
Table 1. GenBank accession numbers for the strains used in this study. The newly isolated strain of Diatrypella macrospora, is shaded. The type species of each genus is indicated as T and ex-type strains are in bold
Species Strains GenBank accession numbers ITS β-tubulin Allocryptovalsa HVFIG02 HQ692573 HQ692524 cryptovalsoidea Allocryptovalsa HVFIG05 HQ692574 HQ692525 cryptovalsoidea Allocryptovalsa elaeidis MFLUCC 15-0707 MN308410 MN340296 Allocryptovalsa polysporaT MFLUCC 17-0364 MF959500 MG334556 Allocryptovalsa rabenhorstii WA08CB HQ692619 HQ692523 Allocryptovalsa rabenhorstii WA07CO HQ692620 HQ692522 Allodiatrype arengaeT MFLUCC 15-0713 MN308411 MN340297 Allodiatrype elaeidicola MFLUCC 15-0737a MN308415 MN340299 Allodiatrype elaeidicola MFLUCC 15-0737b MN308416 - Allodiatrype elaeidis MFLUCC 15-0708a MN308412 MN340298 Allodiatrype elaeidis MFLUCC 15-0708b MN308413 - Allodiatrype thailandica MFLUCC 14-1210 KU315392 - Allodiatrype thailandica MFLUCC 15-0711 MN308414 - Anthostoma decipiensT IPV-FW349 AM399021 - Anthostoma decipiensT JL567 JN975370 JN975407 Cryptosphaeria eunomiaT C1C, CBS 216.87 AJ302417 - Cryptosphaeria eunomiaT C5C, CBS 223.87 AJ302421 - Cryptosphaeria ligniota CBS 273.87 KT425233 KT425168 Cryptosphaeria moravica/ CBS 244.87 HM164735 HM164769 Eutypa petrakii Cryptosphaeria pullmanensis ATCC 52655 KT425235 KT425170 Cryptosphaeria pullmanensis HBPF24 KT425202 GQ294014 Cryptosphaeria subcutanea CBS 240.87 KT425232 KT425167 Cryptosphaeria subcutanea DSUB100A KT425189 KT425124 Cryptovalsa ampelina A001 GQ293901 GQ293972 Cryptovalsa ampelina DRO101 GQ293902 GQ293982 Diatrype brunneospora CNP01 HM581946 HQ692478 Diatrype bullata UCDDCh400 DQ006946 DQ007002 Diatrype bullata D6C, CBS 215.87 AJ302422 - Diatrype decorticata 1056 KU320621 - Diatrype disciformisT D21C, CBS 205.87 AJ302437 - Diatrype disciformisT IRAN 2347C KR605644 KY352434 Diatrype enteroxantha HUEFS155114 KM396617 KT003700 Diatrype enteroxantha HUEFS155116 KM396618 KT022236 Diatrype macowaniana D15C, CBS 214.87 AJ302431 - Diatrype mangrovei MFLUCC 17-0412 MH304407 - Diatrype mangrovei MFLUCC 17-0391 MH304408 - Diatrype mangrovei MFLUCC 17-0394 MH304409 - Diatrype oregonensis CA117 GQ293934 GQ293996 Diatrype oregonensis DPL200 GQ293940 GQ293999 Diatrype palmicola MFLUCC 11-0018 KP744439 - Diatrype palmicola MFLUCC 11-0020 KP744438 - Diatrype polycocca D16C, CBS 213.87 AJ302432 - Diatrype prominens ATCC: MYA-4410 FJ430594 - Diatrype prominens SBen212 KU721868 - Diatrype sp. H2/4b MG020309 - Diatrype sp. H3/2b MG020294 - Diatrype sp. H2/5c MG020292 - Diatrype spilomea D17C AJ302433 - Diatrype stigma DCASH200 GQ293947 GQ294003 Diatrype stigma UCD23-Oe JX515704 JX515670 Diatrype undulata D20C, CBS 271.87 AJ302436 - Diatrype undulata Olrim324 AY354239 - Diatrype whitmanensis CDB011 GQ293954 GQ294010 Diatrype whitmanensis DCHES100 GQ293951 GQ294008 Diatrypella atlantica HUEFS 136873 KM396614 KR259647 Diatrypella atlantica HUEFS 194228 KM396615 KR363998 Diatrypella banksiae CPC 29118 KY173402 - Diatrypella banksiae CPC 29054 KY173401 - Diatrypella cephalanthi CBS 161.32 MH855258 - Diatrypella delonicis MFLUCC 15-1014 MH812994 MH847790 Diatrypella delonicis MFLU 16-1032 MH812995 MH847791 Diatrypella elaeidis MFLUCC 15-0279 MN308417 MN340300 Diatrypella favacea Isolate 380 KU320616 - Diatrypella favacea CBS 198.49 MH856491 - Diatrypella frostii UFMGCB 1917 HQ377280 - Diatrypella heveae MFLUCC 17-0368 MF959501 MG334557 Diatrypella heveae MFLUCC 15-0274 MN308418 MN340301 Diatrypella iranensis IRAN 2280C KDQ18 KM245033 KY352429 Diatrypella macrospora IRAN 2344C KDQ15 KR605648 KY352430 Diatrypella macrospora MFLUCC 21-0010 MW647094 MW677962 Diatrypella major Isolate 1058 KU320613 - Diatrypella prominens DL28A, ATCC 64182 AJ302442 - Diatrypella pulvinata H048 FR715523 FR715495 Diatrypella quercina DL30M AJ302444 - Diatrypella quercina CBS 108.18 MH854666 - Diatrypella sp. C6 KX611072 - Diatrypella sp. ENQ55 KX828138 KY352431 Diatrypella sp. MNQ75B KX828158 KY352432 Diatrypella tectonae MFLUCC 12-0172a KY283084 - Diatrypella tectonae MFLUCC 12-0172b KY283085 KY421043 Diatrypella verruciformisT UCROK1467 JX144793 JX174093 Diatrypella verruciformisT UCROK754 JX144783 JX174083 Diatrypella vulgaris HVFRA02 HQ692591 HQ692503 Diatrypella vulgaris HVGRF03 HQ692590 HQ692502 Diatrypella yunnanensis VT01 MN653008 - Eutypa armeniacae ATCC 28120 DQ006948 DQ006975 Eutypa astroidea E49C, CBS 292.87 AJ302458 DQ006966 Eutypa flavovirens E48C, CBS 272.87 AJ302457 DQ006959 Eutypa laevata E40C CBS 291.87 AJ302449 - Eutypa lataT CBS 290.87 HM164736 HM164770 Eutypa lataT EP18 HQ692611 HQ692501 Eutypa lataT RGA01 HQ692614 HQ692497 Eutypa lejoplaca CBS 248.87 DQ006922 DQ006974 Eutypa leptoplaca CBS 287.87 DQ006924 DQ006961 Eutypa maura CBS 219.87 DQ006926 DQ006967 Eutypa microasca BAFC 51550 KF964566 KF964572 Eutypa sparsa 3802 3b AY684220 AY684201 Eutypella cerviculataT EL59C AJ302468 - Eutypella cerviculataT M68 JF340269 - Eutypella leprosa EL54C, CBS 276.87 AJ302463 - Eutypella leprosa Isolate 60 KU320622 - Eutypella microtheca ADEL200 HQ692559 HQ692527 Eutypella microtheca BCMX01 KC405563 KC405560 Eutypella parasitica CBS 210.39 DQ118966 - Eutypella semicircularis MP4669 JQ517314 - Halodiatrype avicenniae MFLUCC 15-0953 KX573916 KX573931 Halodiatrype salinicolaT MFLUCC 15-1277 KX573915 KX573932 Kretzschmaria deusta CBS 826.72 KU683767 KU684190 Monosporascus cannonballusT CMM3646 JX971617 - Monosporascus ATCC 26931 FJ430598 - cannonballusT Neoeutypella baoshanensisT EL51C, CBS 274.87 AJ302460 - Neoeutypella baoshanensisT HMAS 255436 NR_164038 MH822888 Paraeutypella citricola HVGRF01 HQ692579 HQ692512 Paraeutypella citricola HVVIT07 HQ692589 HQ692521 Paraeutypella vitis UCD2291AR HQ288224 HQ288303 Paraeutypella vitis UCD2428TX FJ790851 GU294726 Pedumispora rhizophoraeT BCC44877 KJ888853 - Pedumispora rhizophoraeT BCC44878 KJ888854 - Peroneutypa alsophila EL58C, CBS 250.87 AJ302467 - Peroneutypa comosa BAFC 393 KF964568 - Peroneutypa curvispora HUEFS 136877 KM396641 - Peroneutypa diminutiasca MFLUCC 17-2144 MG873479 - Peroneutypa diminutispora HUEFS 192196 KM396647 - Peroneutypa kochiana EL53M AJ302462 - Peroneutypa longiasca MFLUCC 17-0371 MF959502 MG334558 Peroneutypa mackenziei MFLUCC 16-0072 KY283083 KY706363 Peroneutypa mangrovei NFCCI-4246 MG844286 MH094409 Peroneutypa rubiformis MFLUCC 17-2142 MG873477 - Peroneutypa scoparia DFMAL100 GQ293962 GQ294029 Peroneutypa scoparia IRAN 2345C KR605646 KY352452 Quaternaria quaternata CBS 278.87 AJ302469 - Quaternaria quaternata IRAN 2348C KR605645 KY352464 Xylaria hypoxylonT CBS 122.620 AM993141 KX271279
-
Phylogenetic analyses
Phylogenetic analyses of a combined ITS and β-tubulin sequence dataset comprised 131 ingroup taxa and two outgroup taxa, namely Kretzschmaria deusta and Xylaria hypoxylon (Xylariaceae). The combined matrix contained 1240 nucleotide sites (ITS: 1-515; β-tubulin: 516-1240). The ML analysis yielded a best-scoring tree with a final ML optimization likelihood value of -18323.132499. The matrix had 887 distinct alignment patterns, with 38.83% of gaps and completely undetermined characters. The ML and BI analyses yielded trees with similar topologies. The clades recovered in the phylogenetic tree are similar in topologies to previous studies (Acero et al. 2004, Dayarathne et al. 2020, Konta et al. 2020). Diatrypella species clustered in five distinct clades as A1, A2, B, C1 and D1 (Fig. 1). Our newly sequenced strain grouped separately from the type species Diatrypella verruciformis in clade D1. It clustered with the type of D. macrospora (IRAN 2344C) and with Diatrype sp. (H2/4b) with statistical support, MLBS 84%, BYPP 1.00 (Fig. 1).
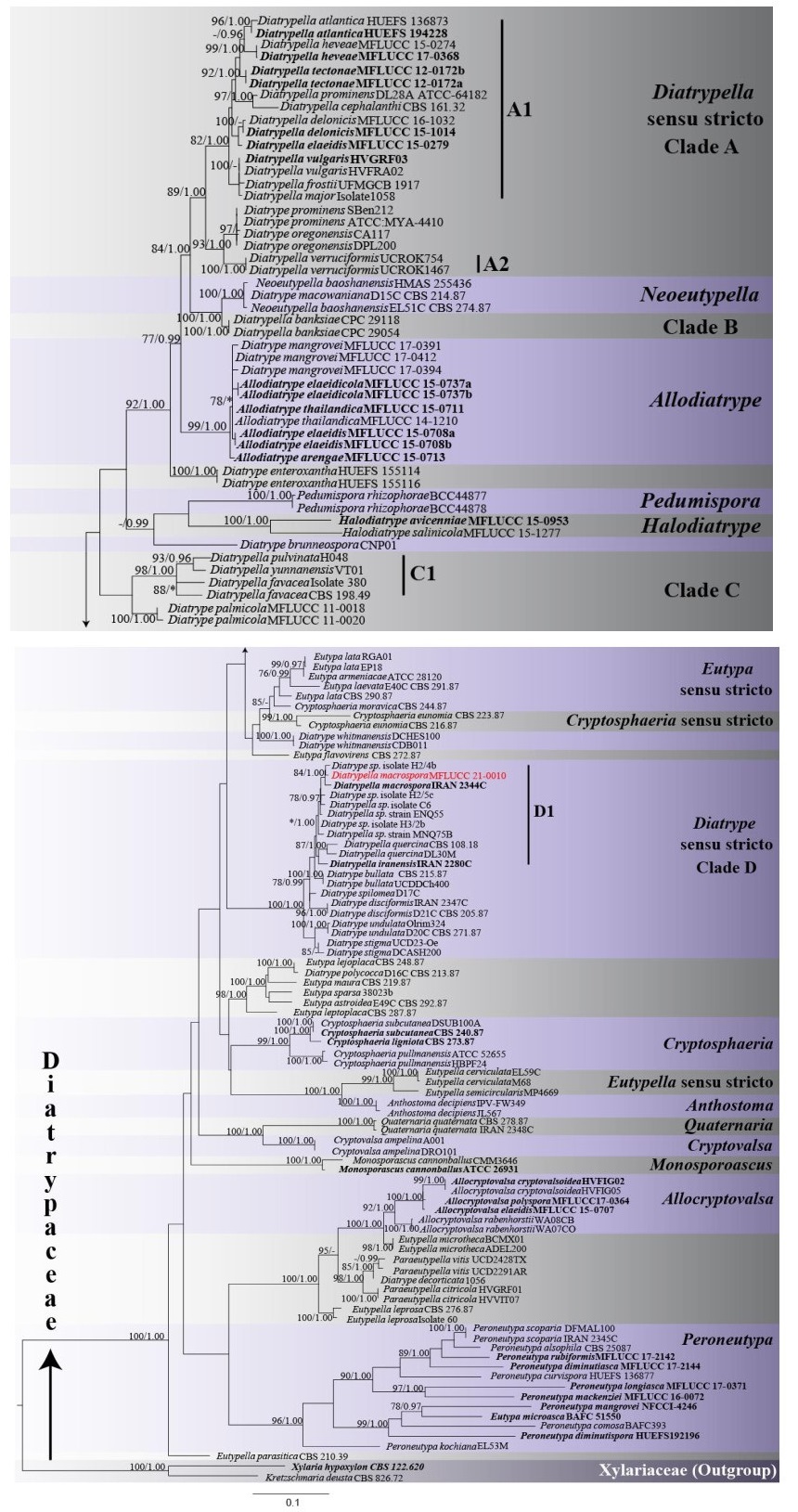
Figure 1. Maximum likelihood phylogenetic tree generated from combined ITS and β-tubulin sequence data of 131 Diatrypaceae taxa and 1240 sites. For each node maximum likelihood bootstrap support values (MLBS) are given first, followed by Bayesian posterior probabilities (BYPP). ML bootstrap support values ≥ 75% and BYPP ≥ 0.95 are indicated at the nodes. Lower values are indicated by a dash (-). Nodes that were not recovered are indicated by asterisks (*). The new isolate is in red bold font. Ex-type sequences are in black bold font. The tree is rooted to Kretzschmaria deusta (CBS 826.72) and Xylaria hypoxylon (CBS 122.620).
Taxonomy
Diatrypella macrospora Mehrabi, Hemmati, Vasilyeva & Trouillas, Phytotaxa 252(1): 47 (2016)
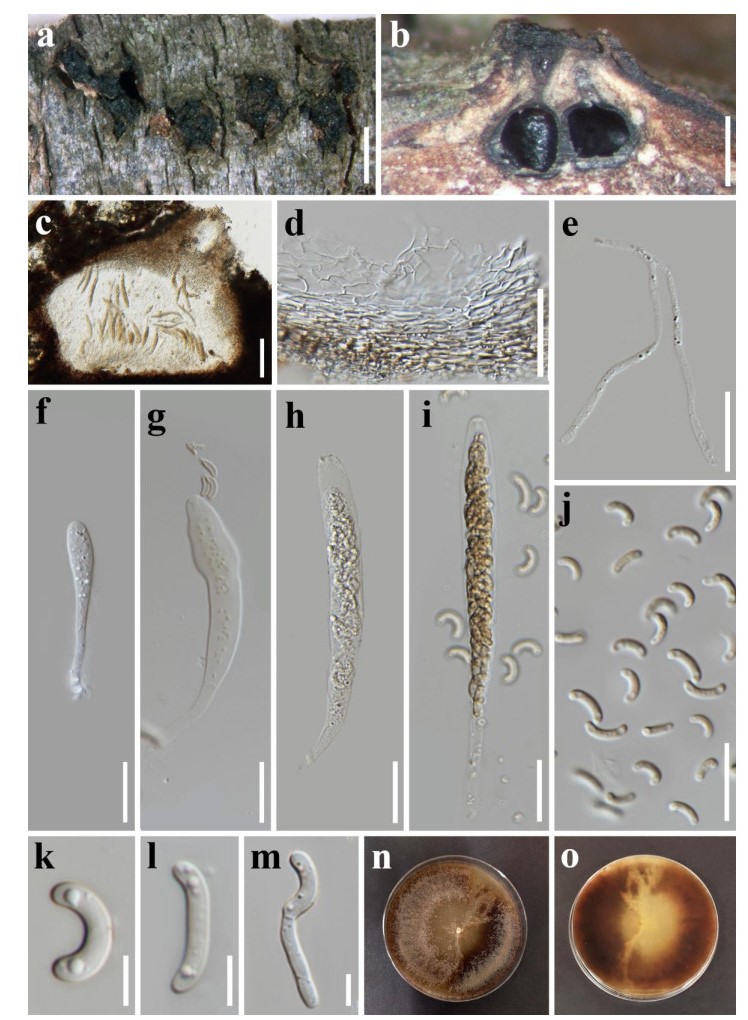
Figure 2. Diatrypella macrospora (MFLU 19-2401, new record). a Stromata erupting from bark. b Longitudinal section through stroma. c Section through a perithecium. d Peridium. e Paraphyses. f-i Asci. j-l Ascospores. m Germinated ascospore. n, o Top and bottom view of culture. Scale bars: a = 2 mm, b = 500 μm, c = 100 μm, d = 50 μm, e-j = 20 μm, k-m = 5 μm.
Index Fungorum number: IF813001; Facesoffungi number: FoF01891
Saprobic on dead land branch of Quercus cerris. Sexual morph: Stromata 1.2-2.8 mm (x = 1.8 mm, n = 10) wide, discoid to irregular, scattered or aggregated, immersed to erumpent arising from cracks in the bark, black, separated from host tissue by a black zone, perithecia arranged in groups of 3- 8, entostroma well-developed, white to yellow to brown. Perithecia 260-490 µm (x = 375 µm, n = 10) in diameter, globoid to flask-shaped, sometimes deformed by compression. Perithecial neck about 110- 350 μm (x = 240 µm, n = 10), ostiolar canals sulcate, compressed, converge together at the apex, black, ostiole brown, opening separately through host bark, periphysate. Peridium composed of light brown to brown, somewhat flattened cells of textura angularis, becoming hyaline towards the inner region. Paraphyses elongate, filiform, aseptate, unbranched. Asci spore-bearing part 72-120 µm × 8-14.5 µm (x = 94 × 11 µm, n = 20), basal part, filiform, 24-53 µm (x = 40 µm, n = 20), polysporous, unitunicate, elongate, cylindrical to clavate, obtuse apex, with a J-apical ring. Ascospores 7.5-12 μm × 2-3.1 µm (x = 10 × 2.5 µm, n = 50), allantoid, subhyaline, yellowish in mass, aseptate, usually bi-guttulate, thin, smooth-walled. Asexual morph: For morphological description see Mehrabi et al. (2016).
Culture characteristics - Colonies on MEA reaching 75 mm diam. after 2 weeks at room temperature. Colonies circular, slightly dense, flat, with fimbriate margin, white to light brown to black from above, similar colour from below.
Material examined - Italy, Province of Forlì-Cesena, Forlì, Farazzano, on a dead land branch of Quercus cerris (Fagaceae), 22 July 2019, E. Camporesi, IT 4432 (MFLU 19-2401, new record, dried culture), living culture MFLUCC 21-0010.
GenBank accession numbers - ITS: MW647094, β-tubulin: MW677962
Notes - Our new collection of Diatrypella macrospora resembles the holotype (IRAN 2344C) as it has subhyaline and allantoid ascospores, elongate asci, overlap in the number of perithecia as well as sizes of stromata, perithecia and asci (Table 2) (Mehrabi et al. 2016). However, our collection differs from D. macrospora (IRAN 2344C) in having smaller ascospores which are considerably more curved and shorter perithecial necks. Based on curvature and spore size, D. macrospora from our collection shows more resemblance with D. quercina (Croxall 1950). In the combined ITS and β-tubulin phylogeny, our new isolate MFLUCC 21-0010 clusters with D. macrospora (IRAN 2344C) and with Diatrype sp. (H2/4b), with high statistical support (MLBS 84%, BYPP 1.00) (Fig. 1, Clade D, D1). There are 2/510 (0.39%) and 9/349 (2.28%) base pair differences between our isolate and D. macrospora (IRAN 2344C) for ITS and β-tubulin sequences respectively. For the ITS sequence of our isolate and Diatrype sp. (H2/4b) there are 2/510 (0.39%) base pair differences. The two strains of Diatrypella macrospora formed a clade with D. iranensis, D. sp. (C6, ENQ55 and MNQ75B), D. quercina, Diatrype bullata, D. disciformis, D. sp. (H2/5c and H3/2b), and D. spilomea, with 78% MLBS and 0.99 BYPP statistical support (Fig. 1, Clade D). Previously, Diatrypella macrospora had been reported only in Iran on Quercus brantii. Our new isolate represents a new host and geographical record of D. macrospora on Quercus cerris (Fagaceae) in Italy.
Table 2. Comparative morphology of Diatrypella macrospora and its relative species
Species Shape of stromata Entostroma colour Perithecial neck (μm) Asci Ascospore Shape, colour, length × width (µm) Reference Shape Number of spores Length (p. sp. µm) Width (µm) Diatrype bullata (representative strain) ovoid white short neck cylindrical 8-spored 25-35 5-7 allantoid, pale yellow, 5-7.5 × 1.2 Rappaz (1987), Vasilyeva & Ma (2014) Diatrype disciformis (reference specimen) orbicular, disc-like yellowish white - cylindrical 8-spored 30-40 5-6 allantoid, pale yellow, 5-9× 1.5-2 Senanayake et al. (2015) Diatrype spilomea (representative strain) effuse white short neck - 8-spored 20-30 3-6 allantoid, pale yellow, 4.5-7 × 1-1.2 Rappaz (1987) Diatrype stigma (representative strain) effuse white short neck cylindrical 8-spored 25-50 5-6 allantoid, pale yellow, 5.8-10.5 × 1.2-2 Rappaz (1987), Vasilyeva & Ma (2014) Diatrype undulata (representative strain) effuse white short neck cylindrical 8-spored 25-40 4-7 allantoid, pale yellow, 5-8 × 1.2-1.8 Rappaz (1987), Vasilyeva & Ma (2014) Diatrypella hevea (Holotype) rounded to irregular white - clavate to cylindric-clavate multispored 80-113 10-21 hyaline to pale yellowish to pale brown oblong to allantoid, aseptate, slightly curved, 5–9 × 1–3 Senwanna et al. (2017) Diatrypella iranensis (Holotype) circular to ovoid whitish yellow - elongate, subcylindrical to clavate multispored - 6-9 allantoid, subhyaline 6-7 × 1-1.3 Mehrabi et al. (2015) Diatrypella macrospora (Holotype) circular white to yellow to light brown relatively long neck, 200–500, converge together elongate, more or less cylindrical 8-spored 110-150 10-15 allantoid, subhyaline 12-20 × 1.7-3 Mehrabi et al. (2016) Diatrypella macrospora (representative strain) discoid to irregular in shape white to, yellow to brown short neck, 110–350, converge together elongate, cylindrical to clavate multispored 72-120 8-14.5 allantoid, strongly curved, subhyaline 7.5–12 × 2–3.1 This study Diatrypella quercina (representative strain) Subregula rrounded or angular - - cylindrical to clavate multispored 80-120 10-12 allantoid, strongly curved 8–12 × 2–3 Croxall (1950), Saccardo (1882) Diatrypella tectonea circular to irregular white to short neck clavate multispored 100-128 15.5-21.5 yellowish to brown, ellipsoidal to cylindrical or elongate-allantoid, 7–9 × 2–2.5 Shang et al. (2017) Diatrypella verruciformis (representative strain) circular, subconical white -
converge togetherspindle-shaped multispored 100-132 11-11.5 allantoid, moderately or variously curved subolivaceous 6–8 × 1–2 Glawe & Rogers (1984)
-
In this study, we identify our collection as Diatrypella macrospora because it is morphologically similar to the holotype, has a short phylogenetic distance, and little ITS base pair differences with the type strain. It also occurs on the same host genus as the holotype. Previously, D. macrospora had only been reported on Quercus brantii in Iran (Mehrabi et al. 2016). Our collection occurs on Quercus cerris, commonly known as Turkey oak. The tree is widespread in the Italian Peninsula (Taffetani et al. 2012) and it is similar to other oak trees but its wood is more prone to cracking and splitting (Vidrinskas & Deveikis 2016). Nonetheless, there are some differences between our strain and the type specimen. The observed differences in morphology may be attributed to differences in host and geographical location. Alternatively, the morphological distinctions along with differences in the nucleotide sequence of β-tubulin also indicate the possibility of a new species rather than a new strain of D. macrospora. However, there is only a single collection of D. macrospora (as well as D. iranensis) and it is therefore difficult to determine species boundaries given the lack of data. Ideally, several differences in more than one locus are required to introduce a new species. With the data currently available, we take a conservative approach and list our isolate as a new record of D. macrospora.
Our analysis also provides insights into Diatrypella. The phylogenetic tree (Fig. 1) clearly shows that the genus is not monophyletic, confirming previous studies (Acero et al. 2004). Diatrypella sequences formed five distinct clades in the tree (Clades A1, A2, B, C1, D1). The newly identified strain along with D. macrospora (IRAN 2344C) formed a clade with D. iranensis and D. quercina, both of which are closely associated with oak trees (Croxall 1950, Farr & Rossman 2020). Further collections are required to determine host-specificity of Diatrypella species in Clade D1. In the phylogenetic tree, the above three species group together in the same clade, distant from the type species, D. verruciformis (Fig. 1), a consistent finding across studies (Mehrabi et al. 2015, 2016, 2019, Konta et al. 2020). Diatrypella verruciformis clusters as sister to Diatrype sequences separately from other Diatrypella. Diatrypella iranensis, D. macrospora and D. quercina could be considered as part of Diatrype, since they form a strongly supported clade with the type D. disciformis. This placement is congruent with previous studies (Acero et al. 2004, Mehrabi et al. 2019, Konta et al. 2020). In these investigations, the three species are treated as part of Diatrype. However, the D. disciformis sequence data used in these studies are not from ex-type strains, thus they might be misidentified. Senanayake et al. (2015) proposed a reference specimen for D. disciformis (MFLU 15-0722, MFLUCC 15-0538), for which LSU and ITS sequences are available. However, these data were not included in the present analysis. In fact, only a few taxa in our dataset have available LSU sequences. Moreover, the ITS sequence for D. disciformis (MFLUCC 15-0538) appears to be problematic. When the strain was subjected to BLAST search, the closest match were members of Nectriaceae, a different family. Therefore, the taxonomic placement of the three Diatrypella species cannot be conclusively confirmed based on molecular data. Similarly, D. favacea, D. pulvinata, D. yunnanensis also form a distinct clade (Clade C1). Clade C1 and D1 could actually represent two distinct and novel genera. However, it is difficult to separate them with certainty from Diatrypella, since there is no sequence from the ex-type of D. verruciformis.
Transfer of D. iranensis, D. macrospora and D. quercina to Diatrype would need revision of the generic concepts or a new genus should be erected. Cesati & De Notaris (1863) separated Diatrypella and Diatrype solely based on the number of spores per ascus (Croxall 1950, Acero et al. 2004). In early taxonomic studies, it was recognized that, although convenient, this classification system is likely to be artificial and not a good reflection of evolutionary history of the two genera (Glawe & Rogers 1984, Rappaz 1987). In fact, phylogenetic analyses show that number of spores per ascus is highly variable throughout the evolution of Diatrypaceae and multispored asci appeared several times independently (Dayarathne et al. 2020, Acero et al. 2004). This explains the mixed clades (Clade A, C, D) comprising both Diatrypella and Diatrype species recovered in this study and previous studies (Acero et al. 2004, de Almeida et al. 2016, Dayarathne et al. 2020, Konta et al. 2020). Apart from the number of spores per ascus other unique characteristics should be used to delineate between genera (Carmarán et al. 2006). However, from the data available (Table 2) there is no unique common characteristic separating Diatrypella species in Clade C1 from those in Clade A.
As mentioned by Acero et al. (2004), several early taxonomists have highlighted the morphological distinction between D. quercina and other Diatrypella species (Acero et al. 2004). Specifically, although the species has polysporous asci it also has a well-developed ectostroma, which is unusual of Diatrypella, but common in Diatrype (Wehmeyer 1926). Diatrypella macrospora (MFLUCC 21-0010), which groups in the same clade as D. quercina, also displays a higher degree of ectostromatic development. No details were given for the ectostroma of D. iranensis and D. macrospora (IRAN 2344C). Thus, even though ectostroma could be used as a taxonomic character this cannot be conclusively determined due to lack of information. This could be because the ectostroma is normally not used as taxonomic characteristic for herbarium as it is present only on immature specimens (Rappaz 1987). Also, some taxonomists argue that microscopic characteristics are more appropriate for delineating diatrypaceous taxa as compared to the more variable macroscopic characters (de Almeida et al. 2016).
Another difference observed by Croxall (1950) in D. quercina is its strongly curved ascospores. However, curvature in ascospore appears only as a difference between species rather than between genera (Table 2). Molecular analysis confirmed the distinction of D. quercina from other Diatrypella species (Acero et al. 2004). The morphological characteristics related to these molecular differences are not clear. A summary of recent morphological descriptions of species in Clade D is also provided by Thiyagaraja et al. (2019). The data presented is coherent with that reviewed in our study except for the colour of the entostroma which seems to have been mixed with the colour of stromata. Still, the additional details do not provide insights into a more reliable classification. From the morphological descriptions available, D. iranensis, D. macrospora and D. quercina show no clear common feature that separate them from other Diatrypella species or explain their phylogenetic position among Diatrype species (Table 2). Collectively, these results suggest that the currently used morphological features have a high degree of overlap and their taxonomic value might need to be reconsidered.
Missing data, inaccessibility of type specimens and inadequate original descriptions make resolving the taxonomic problems mentioned above challenging. The genus concept of Diatrypella and Diatrype as well as the classification of Diatrypaceae as a whole should be reviewed. Sequencing of the reference specimen for Diatrype should be checked and an epitype should be provided for Diatrypella (de Almeida et al. 2016). More collections of diatrypaceous taxa and a combination of reliable molecular and morphological information would greatly aid resolving the taxonomy of Diatrypaceae.
- We would like to thank Mae Fah Luang University and the Mushroom Research Foundation (MRF), Chiang Rai for financial support. We are grateful to Chunfang Liao for sequencing of sample, Dr. Eleni Gentakaki and Dr. Saranyaphat Boonmee for their great assistance. K.D. Hyde expresses his thanks to the Thailand Research Fund ("Impact of climate change on fungal diversity and biogeography in the Greater Mekong Sub-region RDG6130001").
- Copyright: © 2021 by the author(s). This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
| JE Carpouron, SN Wijesinghe, QJ Shang, RH Perera, E Camporesi, KD Hyde. 2021. Diatrypella macrospora, a new host and geographical record from Forlì-Cesena, Italy. Studies in Fungi 6(1):273−287 doi: 10.5943/sif/6/1/18 |












