HTML
-
Citrofortunella microcarpa locally known as "kalamansi" belonging to the family Rutaceae, is one of the most important commercial fruit crops grown in the Philippines and used condiment in almost every famous dish (Morte et al. 2017). It is characterized as small tree with smooth, oblong to broadly lanceolate, and narrowly winged short petioled leaves (Dulay & De Castro 2016). Aside from being a fruit crop, this plant also houses various endophytic microorganisms that are found in wide variety of plant tissues such as seeds, ovules, fruits, stems, roots, leaves, tubers, buds, xylem and bark. Endophytic microorganisms are inhabitants of the internal part of plants, causing apparently no harm to their hosts. It has been also found that some endophytic fungi produce valuable enzymes that are relatively unexplored which are useful to pharmaceutical and agricultural industries producing valuable substances of biotechnological interest (Patel et al. 2013). Enzymes produced by microorganisms are considered as potential biocatalysts for a large number of reactions and are generally regarded as safe and functional at wide range of temperature, pH, salinity or other extreme conditions (Priyadharsini & Dhanasekaran 2015). Thus, this study was conducted to evaluate the enzymatic ability of endophytic fungi isolated from C. microcarpa stem and leaves that would provide information on their diversity and ability to produce fungal enzymes such as amylase, cellulose, protease and laccase that is of biotechnological interest.
-
Citrofortunella microcarpa stem and leaves were collected from calamansi plantations in Mayapyap Sur, Cabanatuan City, Nueva Ecija (15°31'05.0"N 120°57'48.9"E). Matured and healthy leaves and stem were cleaned of adhering debris and surface sterilized using 70% ethanol for 1 minute followed by sequential 1-minute washing with solutions containing 1% sodium hypochlorite, 70% ethanol and sterile distilled water and allowed to surface dry on filter paper. After proper drying, approximately 1x1 cm of sterilized leaf and 1cm length of stem were inoculated in a previously plated potato dextrose agar (PDA) and incubated at room temperature for 5 days. Then, individual colonies were inoculated onto new PDA plates and repetitive re-plating of the fungal colonies was continued until pure cultures were obtained (Patel et al. 2013). Pure cultures of the identified isolates were deposited at Biodiversity Conservation Laboratory, Department of Biological Sciences, College of Science, Central Luzon State University, Science City of Munoz, Nueva Ecija, Philippines, 3120. Facesoffungi numbers were registered for the fungal cultures as per Jayasiri et al. (2015).
Cultural and morphological characterization of fungal species
-
Cultural characteristics such as colony appearances, mycelial textures and pigmentations on both obverse and reverse sides of PDA plates were observed after 7 days of incubation at room temperature. Growth rate via colony diameter were measured, initially standardized at 6 mm using a cork borer. Trials were repeated three times and in triplicates (Majid et al. 2015). For microscopic observation, slide culture was used to observe the different micro-morphological characteristics of the isolated fungal endophytes (Prakash & Bhargava 2016).
Molecular identification of fungal endophytes
-
Seven-day old cultures of the different fungal isolates grown on PDA medium on test tubes were sent to the Philippine Genome Center in Quezon City, Manila for DNA extraction and sequencing. DNA extraction was done using CTAB method. Gene was amplified by PCR and components include genomic DNA, Universal ITS1 (5'tccgtaggtgaacctgcgg-3') and ITS4 (5'-tcctccgcttattgatatgc-3') primers, Taq Buffer, DNA Polymerase, and dNTP Mix. Cycling parameters on thermal cycler: 95℃ 5 min; 30 cycles of 95℃ 1 min, 58℃ 45 secs, 70℃ 1 min; 72℃ 10 min; hold at 4℃. Capillary sequencing was carried out on the ABI 3730xl DNA Analyzer using a 50cm 96-capillary array, POP7TM Polymer, and 3730xl. Data Collection Software v3.1 and base calling was done on Sequencing Analysis Software v5.4. Trimming and assembling of sequences was done using Codon Code Aligner V8.0.2. Sequencing was verified by BLASTn and aligned through CLUSTAL W.
Isolation of fungal endophytes
-
Fungal endophytes from C. microcarpa were grown on glucose yeast peptone agar (glucose: 1g; yeast extract: 1g; peptone: 0.5g; agar: 16g; d.w.: 1000ml; pH 6) medium supplemented with 2% starch as substrate. Then, plates inoculated with fungal endophytes were incubated at room temperature for 7 days. After incubation, the plates were flooded with 1% iodine solution. The clear zone formed surrounding the fungal colonies were considered positive for the test (Patel et al. 2013).
Cellulase activity
-
Fungal endophytes from C. microcarpa were grown on yeast peptone agar medium (yeast extract: 1g; peptone: 0.5g; agar: 16g; d.w. 1000ml) supplemented with 0.5% Na-carboxymethyl cellulose (CMC) as enzyme substrate. Then, plates were incubated for 7 days at room temperature. After incubation, the plates were flooded with 0.2% aqueous solution of Congo red and followed by destaining using 1M NaCl solution or 15 min. The clear zone surrounding the colony indicated positive for the test (Patel et al. 2013)
Laccase activity
-
Fungal endophytes from C. microcarpa were grown on glucose yeast peptone agar medium (glucose: 1g; yeast extract: 1g; peptone: 0.5g; agar: 16g; d.w.: 1000ml; pH: 6) amended with 0.005% 1-naphthol as enzyme substrate. Plates were the incubated at room temperature for 7 days. Oxidation of 1-naphthol by laccase, the culture medium changed from clear to blue. The color change indicated positive for the test (Patel et al. 2013).
Protease activity
-
Fungal endophytes from C. microcarpa were grown on glucose yeast peptone medium (glucose: 1g; yeast extract; 1g; peptone; 0.5g; agar; 16g; d.w.; 1000ml; pH: 6) supplemented with 0.4% gelatin as enzyme substrate Then, plates inoculated with fungal endophytes were incubated at room temperature for 7 days. After incubation, saturated aqueous ammonium sulphate was used to flood the plates. The digested area surrounding the colonies appeared clear indicating positive for the test (Patel et al. 2013).
Data analysis
-
The enzymatic index (EI) were determined by dividing the measured colony diameter (Øc) and the formed enzymatic halo diameter (Øh) (EI = Øh/Øc). Samples with computed enzymatic index with 2.0 and higher are considered to have a high rate of enzyme production (Herculano et al. 2011). The halo presence or clear area around the fungal colony were the enzymatic activity indicator evidenced by the enzymes secretion made by fungi through the culture medium (Sharma & Sumbali 2014).
Statistical analysis
-
The enzymatic experiments were laid out on a Completely Randomized Design (CRD). Screening for enzyme activities were analyzed using analysis one-way Analysis of Variance (ANOVA). Comparison among means was done using Duncan multiple range test (DMRT) at 5% level of significance using IBM SPSS Statistics 2019 Ver. 26 (SPSS Inc., Chicago, USA).
Amylase activity
-
A total of 11 fungal endophytes were isolated from stem and leaves of Citrofortunella microcarpa namely; Colletotrichum fructicola, Colletotrichum gloeosporioides, Colletotrichum siamense, Fusarium oxysporum, Lasiodiplodia theobromae, Nigrospora oryzae, Nigrospora rubi, Nodulisporium indicum, Phomopsis azadirachtae, Phyllosticta capitalensis, and an unidentified Pleosporales species. C. fructicola and F. oxysporum were isolated from leaves. N. indicum, L. theobromae, P. azadirachtae, P. capitalensis and the unidentified species were isolated from stem. Meanwhile, N. oryzae, C. siamense, N. rubi and C. gloeosporioides were isolated both from leaves and stem.
To confirm the identity of the fungal endophytes isolated from C. microcarpa, the ITS region was amplified using ITS1 and ITS4 primers and were sequenced. The PCR products of the ITS region in the eleven different fungi were confirmed to be in range of approximately 600-900bp which are characteristics of ITS1 and ITS4 region. Blast analysis revealed that fungal species were identified as C. fructicola FoF 06767 with (99.79%), C. gloeosporioides FoF 09424 (99.79%), C. siamense FoF 03599 (99.57%), F. oxysporum FoF 03824 (98.13%), L. theobromae FoF 00167 (99.02%), N. oryzae FoF 06596 (99.58%), N. rubi FoF 06598 (99.38 %), N. indicum FoF 10588 (99.36%), P. azadirachtae FoF 10589 (98.31%), P. capitalensis FoF 06888 (99.64%), and unidentified species FoF10590 (95.83%), respectively (Table 1.)
Table 1. Identities of the cultured fungi based on BLAST search with NCBI Genbank accessions and Facesoffungi numbers
Isolate No. Facesoffungii Number Species E value Identity Gen Bank Accession No. LF01 FoF 06767 Colletotrichum fructicola 0.00 99.79 % MK041513 LF02 FoF 03824 Fusarium oxysporum 0.00 98.13 % MG543712 STM01 FoF 00167 Lasiodiplodia theobromae 0.00 99.02 % KF814723 STM02 FoF 10588 Nodulisporium indicum 0.00 99.36 % KJB26516 STM03 FoF 10589 Phomopsis azadirachtae 0.00 98.31 % KR056296 STM05 FoF 06888 Phyllosticta capitalensis 0.00 99.64 % MF153383 STM06 FoF10590 Unidentified species 0.00 95.83 % HM751829 STM/LF01 FoF 03599 Colletotrichum siamense 0.00 99.57 % MK569274 STM/LF02 FoF 06596 Nigrospora oryzae 0.00 99.58 % KU360637 STM/LF03 FoF 06598 Nigrospora rubi 0.00 99.38 % KX985948 STM/LF04 FoF 09424 Colletotrichum gloeosporioides 0.00 99.79 % KP900290 LF = Isolated from leaf, STM = Isolated from stem
Cultural and morphological characteristics of fungal endophytes
-
The eleven species of endophytic fungi tested were able to produce amylase (Table 2, Fig. 1). P. capitalensis had the highest computed enzymatic index which is 3.75 followed by the unidentified species and N. oryzae with 1.99 and 1.60, respectively. Computed values were significantly different from each other. Meanwhile, N. indicum, F. oxysporum, L. theobromae, N. rubi, C. siamense and P. azadirachtae all have computed enzymatic index of 1.0 which are significantly different from C. fructicola.
Table 2. Amylase activity of different fungal endophytes after seven days of incubation
Fungal endophytes Amylase Diameter of clear zone (mm) Colony diameter (mm) Enzymatic index (EI) Nodulisporium indicum 85.00a 85.00 1.00d Fusarium oxysporum 85.00a 85.00 1.00d Lasiodiplodia theobromae 85.00a 85.00 1.00d Nigrospora rubi 85.00a 85.00 1.00d Colletotrichum siamense 85.00a 85.00 1.00d Phomopsis azadirachtae 83.47a 83.47 1.00d Colletotrichum fructicola 75.88b 75.96 0.99e Colletotrichum gloeosporioides 65.80c 65.80 1.00d Nigrospora oryzae 44.10d 27.63 1.60c Unidentified species 40.90e 20.53 1.99b Phyllosticta capitalensis 23.50f 6.00 3.75a Values represent the mean measurements of the clear zones indicating amylase activity produced by the fungal endophytes from Citrofortunella microcarpa. Means with the same letter superscript are not significantly different at 5% level of significance using DMRT
***EI greater than 2.00 has high rate of enzyme production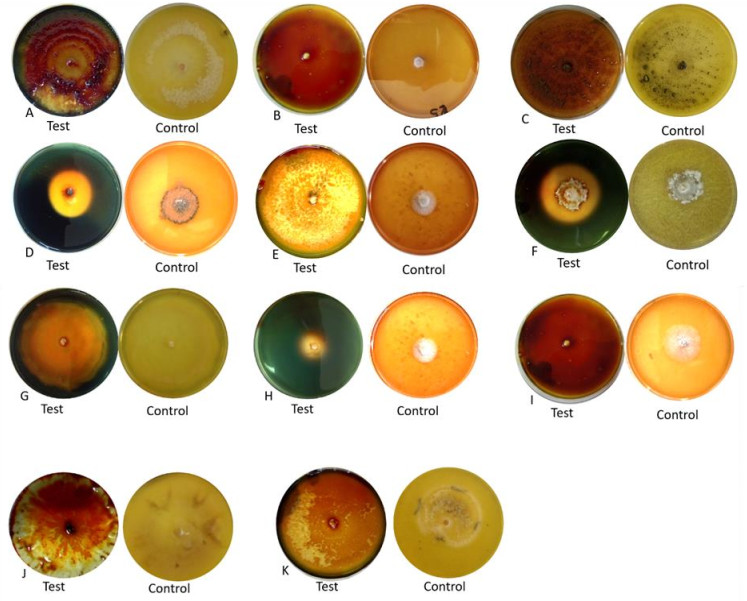
Figure 1. Clear zone produced by different fungal endophytes on amylase activity isolated from C. microcarpa. A Colletotrichum fructicola. B Lasiodiplodia theobromae. C Nodulisporium indicum. D Phyllosticta capitalensis. E Colletotrichum siamense. F Nigrospora oryzae. G Colletotrichum gloeosporioides. H unidentified species. I Fusarium oxysporum. J Nigrospora rubi. K Phomopsis azadirachtae.
Cellulase activity
-
A total of four species of endophytic fungi isolated from C. microcarpa namely L. theobromae, N. oryzae, C. gloeosporioides and unidentified Pleosporales showed cellulose degradation (Table 3, Fig. 2). The largest enzymatic index was unidentified species with an index of 2.02, second was C. gloeosporioides with 1.22 while L. theobromae and N. oryzae both having 1.06 computed enzymatic index respectively. Meanwhile, L. theobromae, and N. oryzae were comparable with each other. Only 25% of the endophytes tested in this study were able to produce cellulose.
Table 3. Cellulase activity of different fungal endophytes after seven days of incubation.
Fungal endophytes Cellulase Diameter of clear zone (mm) Colony diameter (mm) Enzymatic index (EI) Nigrospora oryzae 80.0a 75.60 1.06c Lasiodiplodia theobromae 78.4b 74.20 1.06c Colletotrichum gloeosporioides 74.9c 61.20 1.22b Unidentified species 25.2d 12.70 2.02a Values represent the mean measurements of the clear zones indicating cellulase activity produced by the fungal endophytes from Citrofortunella microcarpa. Means with the same letter superscript are not significantly different at 5% level of significance using DMRT
***EI greater than 2.00 has high rate of enzyme production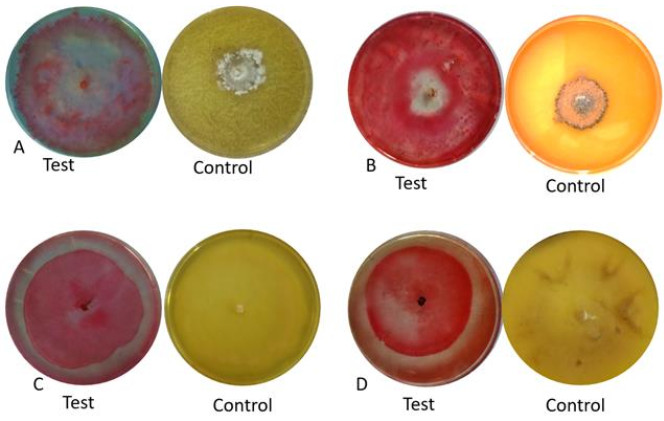
Figure 2. Clear zone produced by different fungal endophytes on cellulase activity isolated from Citrofortunella microcarp. A Nigrospora oryzae. B unidentified Pleosporales. C Colletotrichum gloeosporioides. D Lasiodiplodia theobromae.
Protease Activity
-
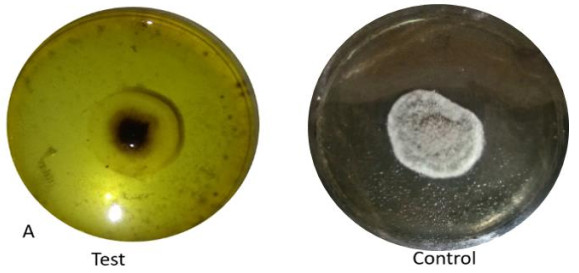
Among the tested organisms, extracellular protease activity was only observed in unidentified Pleosporales with a clear zone of 31.3 mm and computed enzymatic index of 1.47 (Fig. 3).
Laccase Activity
-
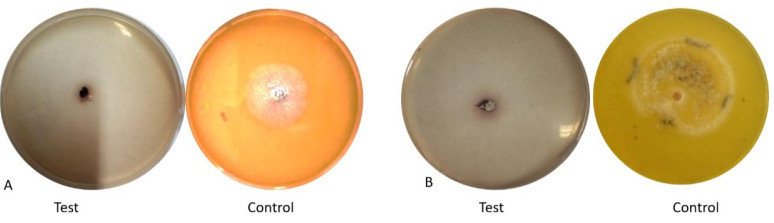
Fusarium oxysporum and Phomopsis azadirachtae were producers of laccase enzyme (Table 4, Fig. 4). Only two endophytic fungi in this study were able to produce laccase.
Table 4. Laccase activity of different isolated fungal endophytes after seven days of incubation
Fungal endophytes Laccase Colletotrichum fructicola - Colletotrichum gloeosporioides - Colletotrichum siamense - Fusarium oxysporum + Lasiodiplodia theobromae - Nigrospora oryzae - Nigrospora rubi - Nodulisporium indicum - Phomopsis azadirachtae + Phyllosticta capitalensis - Unidentified species - (+) presence of color change, (-) absence of color change
Amylase activity
-
The role of endophytic fungi in plant decomposition has been demonstrated over several years, since these fungi are present in the senescent tissue, and they are the first ones to initiate plant decomposition (Wilson 2000). Furthermore, Sun et al. (2011) also indicated that endophytes generally decompose dead host leaves, not only as a single species, but also as communities. In this study, 11 fungal endophytes were isolated and identified from both leaves and stem of Citronella microcarpa. These were three species of Colletotrichum, two species of Nigrospora and species each for the genus Fusarium, Lasiodiplodia, Nodulisporium, Phomopsis, Phyllosticta and unidentified Pleosporales which were tested for their enzyme producing abilities.
Endophytes are fungi that asymptomatically colonize plant tissues during some phase of their life cycle (Saikkonen 2007), but may turn pathogenic during host senescence (Rodriguez et al., 2009). The fungal endophytes isolated and identified were found to be associated from various diseases of Citrus. Colletotrichum, Fusarium, and Lasiodiplodia species were found to be associated with citrus diseases in Europe, Mediterranean and Florida (Yaseen & D'Onghia 2010, Zhang 2014, Guarnaccia et al. 2017a). Also, Phyllosticta and Phomopsis are causal agents of Citrus disease such as the stem-end rot and Citrus Black Spot Disease (Gopal et al. 2013, Guarnaccia et al. 2017b). Meanwhile, Nodulisporium produced antifungal volatile compounds that can control green mold decay on Citrus limon (Suwannarach et al. 2013) while Nigrospora, a ubiquitious endophytes, have been studied for the discovery of novel metabolites (Wang et al. 2017). Describing the niche and ecological role of endophytes are complicated because identical fungal species have been labelled as endophytic or pathogenic depending on the basis of the study and whether the study examines asexual or sexual stage of the fungal species (Ahlholm et al. 2002).
One of the most important enzymes used in biotechnology involved in starch hydrolysis are amylases. Amylases produced by various fungi have vast application in food and medicine production such as baking, brewing, preparation of digestive aids (Couto & Sanromán 2006). As a result of the amylase test, all fungal endophytes isolated and identified were able to degrade starch. Based on the studies of Venkatesagowda et al. (2012) they have found out that Colletotrichum gloeosporioides, Fusarium oxysporum and Lasiodiplodia theobromae produce amylolytic activity.
Cellulolytic microorganisms play an important role in the biosphere by recycling cellulose, the most abundant renewable carbohydrate produced by plants through the mechanism of photosynthesis (Qin et al. 2010). N. oryzae, and the unidentified species were able to show cellulose degredation similar with the results of the previous study by Qin et al. (2010). Moreover, cellulase activity was also detected on C. gloeosporioides and L. theobromae (Toghueo et al. 2017).
Proteases are one of the major enzymes utilized widely as detergents, used in leather, food and pharmaceutical industries and for bioremediation processes (Najafi et al. 2005). Proteolytic enzymes play an important role in fungal physiology and development (Yike 2011).
Laccase is blue copper dependent oxidases which is a main ligninolytic enzyme produced by white rot fungus and catalyzing the oxidation of large numbers of phenolic compound (Poonkuzhali et al. 2011). Fungal laccases have boundless biotechnological functions such as discoloration and detoxification of industrial effluent, bleaching of pulp, phenolics elimination from wine and many others (Yaver et al. 2001). Only two endophytic fungi in this study were able to produce laccase, similar to the results of Sunitha et al. (2013), showing that species of Fusarium and Phomposis showed laccase activity.
- Copyright: © 2021 by the author(s). This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
| RDC Reyes, AM Parayao, KGD Waing. 2021. Identification and evaluation of enzymatic ability of fungal endophytes from Citrofortunella microcarpa (Bunge) Wijnands. Studies in Fungi 6(1):460−468 doi: 10.5943/sif/6/1/35 |