HTML
-
Grapevine is considered as one of the economically important horticultural crops in Ontario. The provincial growers produced 77, 775 tons of grapes with farm value of $111.2 Mil in 2019 (OMAFRA). Some important fungal diseases and pathogens affecting grapevine yield are still poorly known in Canada, including trunk diseases.
The recent studies of grapevine diseases in British Columbia (Okanagan, Similkameen, Fraser valleys, and the Vancouver and Gulf islands) have revealed that the grapevine trunk diseases (GTD) are associated with fungal pathogens of Cadophora, Phaeomoniella, Phaeoacremonium and Togninia genera as well as with the cancer and dieback diseases caused by Botryosphaeria, Diaporthe and Eutypa species. These pathogenic fungi cause the diseases limiting both vineyard longevity and productivity in British Columbia (O'Gorman et al. 2013, Úrbez-Torres et al. 2014). The common dieback symptoms appear as branch stunting, leaf distortion and perennial cankers that can finely result in grapevine decline (Carter 1991, Úrbez-Torres et al. 2009).
One of the grapevine cankers and consequent dieback diseases, Eutypa dieback, is usually associated with Eutypa lata (E. lata) (Rolshausen et al. 2006). However, the recent reports show that different diatrypaceous fungi can cause Eutypa dieback: Eutypa leptoplaca (Trouillas & Gubler 2004), Cryptovalsa ampelina (Luque et al. 2006) and E. vitis (Catal et al. 2007). The pathogen, E. vitis, was not previously reported from Ontario on grapevines as well as on other host plants. However, the fungus has been located in North America (Vasilyeva & Stephenson 2006) and is known in adjacent Michigan State of the USA (Jordan & Schilder 2005). E. vitis and other diatrypaceous fungi have been found to be pathogenic to grapevines in Texas, USA (Úrbez-Torres et al. 2009). It has also reported that the species is associated with branch dieback of Japanese persimmon trees (Diospyros kaki Thunb.) (Firoozjah et al. 2015) and Persian walnut trees (Juglans regia L.) in Iran (Sohrabi at al. 2020) indicating that these tree species as alternative hosts play the role of fungal inoculum's reservoirs that may contribute to the epidemiology of grapevine dieback.
For this reason, we surveyed the local forest stands in the Niagara region of Southern Ontario (Canada) to find any trees and shrubs with symptoms similar to Eutypa dieback. This kind of branch dieback was observed on several tree species (Fagus grandifolia Ehrh., Fraxinus pennsylvanica Marshall and Syringa reticulata (Blume) H. Hara)) in the studied area.
The main aim of this study was to isolate and identify the causative agent of the dieback on the trees growing in the region.
-
The samples of affected branches were collected and single spore isolates were obtained following the method described in Phookamsak et al. (2015). The isolates were plated on malt extract agar (MEA, Seaweed Solution Laboratories, USA; 35 g/l litre of sterile water) and incubated at room temperature (20–25ºC) in the dark. Germinating ascospores were aseptically transferred onto fresh MEA plates. Microscopic examinations, photomicrographs and measurements of 50 randomly selected spores from 3 different samples were taken with an AmScope B120C-E5 stereo microscope and AmScope SE306R-PZ dissecting microscope with a 5 MP digital AmScope camera MD500 supplied with AmScopeX software for Windows v4.8.15934 (AmScope, Irvine, USA). The voucher specimens were deposited into the Herbarium of the Nature Research Centre (BILAS), Institute of Botany, Vilnius, Lithuania. Facesoffungi number was registered for E. vitis as mentioned in Jayasiri et al. (2015).
DNA was extracted by performing a crude NaOH lysis on each colony (EI-13-ON, EI-109-ON) picked. The internal transcribed spacer region (ITS) was amplified using primers ITS1/ITS4 (White et al. 1990) followed by enzymatic cleanup and sequenced using the BigDye Terminator v3.1 on an Applied Biosystems 3730XL (Applied Biosystems, Foster City, California, United States) high throughput capillary sequencer according to the GENEWIZ SOP at GENEWIZ, Inc. (South Plainfield, New Jersey, United States).
The newly obtained and retrieved based the recently published studies on Diatrypaceae (Trouillas et al. 2010, 2011, Grassi et al. 2014, Thiyagaraja et al. 2019) ITS sequences (Table 1) were aligned in CLUSTAL-X2 (Thompson et al. 1997) and edited manually in MEGA-X (Kumar et al. 2018). Some characters were trimmed from both ends of the alignment to approximate the size of the obtained sequences to those included in the dataset. Phylogenetic analysis of the aligned data was performed employing maximum-likelihood (ML) and Bayesian inference (BI) analysis. Maximum likelihood analysis was executed by Randomized accelerated maximum likelihood (RAxML) method using raxmlGUI v. 2.0. A 3-parameter model with unequal base frequencies (TPM3uf) was applied with discrete gamma distribution complemented for each substitution model with four rate classes (Silvestro & Michalak 2011). One thousand bootstrap analysis and searches for the best-scoring ML tree were executed (Stamatakis et al. 2008). Bayesian posterior probabilities (BP) were defined by Markov Chain Monte Carlo sampling (MCMC) with GTR model in MrBayes v.3.2.7 (Huelsenbeck & Ronquist 2001). Six simultaneous Markov Chains were run for 100, 000 generations. Trees were sampled every 100 generation. The first 1000 trees were discarded, and the remaining trees were used for calculating posterior probabilities in the majority rule consensus tree.
Table 1. Fungal taxa with GenBank accession numbers of ITS sequences used in phylogenetic analyses. The strains and GenBank accession numbers of newly generated sequences from this study are in black bold
Taxon Strain GenBank Accession Numbers of ITS Sequences Allocryptovalsa rabenhorstii WA07CO HQ692620 Allocryptovalsa rabenhorstii WA08CB HQ692619 Anthostoma decipiens IPVFW349 AM399021 Anthostoma decipiens JL567 JN975370 Cryptosphaeria ligniota CBS 273.87 KT425233 Cryptosphaeria pullmanensis HBPF24 KT425202 Cryptosphaeria pullmanensis ATCC 52655 KT425235 Cryptosphaeria subcutanea DSUB100A KT425189 Cryptosphaeria subcutanea CBS 240.87 KT425232 Cryptovalsa ampelina A001 GQ293901 Cryptovalsa ampelina DRO101 GQ293902 Eutypella citricola HVVIT07 HQ692579 Eutypella citricola HVGRF01 HQ692589 Eutypella vitis UCD2484TX FJ790854 Eutypella vitis 59 KU320620 Eutypella vitis EI-13-ON MW145141 Eutypella vitis EI-109-ON MW145154 Peroneutypa alsophila EL58C AJ302467 Peroneutypa diminutispora MFLUCC 17-2144 MG873479 Peroneutypa kochiana EL53M AJ302462 Peroneutypa rubiformis MFLUCC 17-2142 MG873477 Peroneutypa scoparia MFLUCC 11-0478 KU940151 Quaternaria quaternate GNF13 KR605645 Quaternaria quaternate CBS 278.87 AJ302469 Xylaria hypoxylon CBS 122620 AM993141
-
Eutypella vitis (Schwein.) Ellis & Everh., N. Amer. Pyren. (Newfield): 490 (1892) Fig. 1
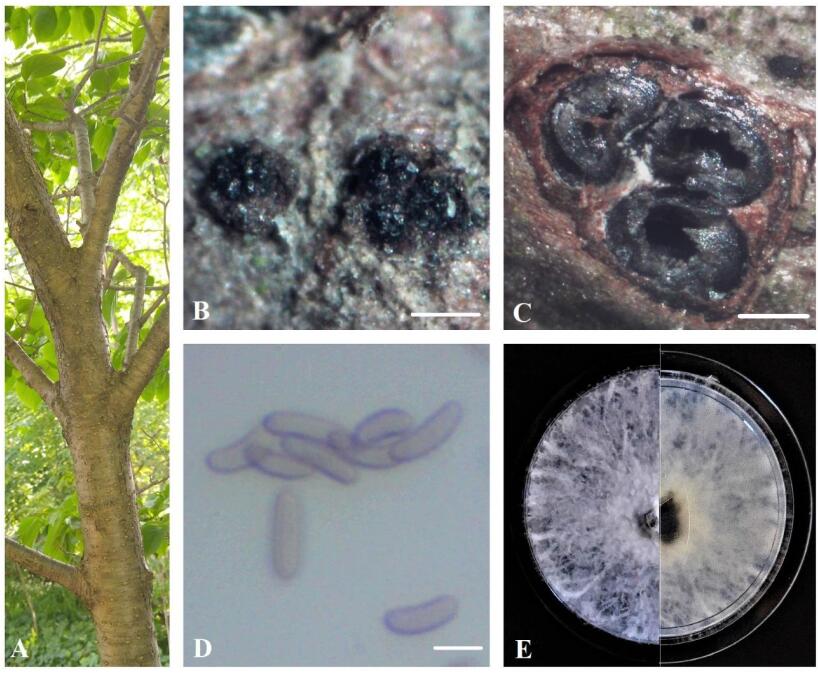
Figure 1. A Eutypa dieback symptoms on Syringa reticulata. B Stromata. C Transverse section through stroma. D Ascospores. E Seven-day-old culture on MEA. Scale bars: B = 0.5 mm, C = 100 μm, D = 5 μm.
Index Fungorum number: IF 123583; Facesoffungi number: FoF 09536
Sexual morph: Ascostromata 0.7-1.3 mm diam., black, superficial, subglobose or ellipsoidal, carbonaceous. Ascomata 120–350 μm diam., perithecial, black, clustered, semi-immersed, ovoid, with 2–4 locules. Asci 35–45 × 8–15 μm (n = 10), 8-spored, thin-walled, cylindric or slightly clavate, some rounded at apex. Ascospores 8.5–10 × 2–3.0 μm (x = 9.2 × 2.6 μm, n = 50), numerous, light brown to brown, allantoid, aseptate, slightly curved, smooth-walled, some with guttules.
Culture characteristics – Colonies on MEA reach 5 cm diam. after 7 days of growth at room temperature in the dark, irregular, slightly raised, cottony, white, becoming beige, margin rough.
Material examined – Canada, Ontario, Niagara Region: EI-13-ON, on dying branches of Fagus grandifolia, 43°11'59.9"N 79°13'37.6"W, 14.05.2020 (BILAS 51392); EI-21-ON, on dying twigs of Syringa reticulata, 43°13'12.3"N 79°13'35.7"W, 26.05.2020 (BILAS 51393); EI-109-ON, on dying branches of Fraxinus pennsylvanica, 43°05'55.2"N 79°16'15.5"W, 22.06.2020 (BILAS 51394).
Phylogenetic analyses
-
The dataset included the representatives of the family Diatrypaceae and the strains obtained from the study. Based on BLASTn search of the NCBI database using ITS sequences for both EI-13-ON (MW145141) and EI-109-ON (MW145154), the top hits were E. vitis specimens collected in USA [GenBank KU320620; Identities = 579/581(99%), Gaps = 0/581(0%)], [GenBank AY462566; Identities = 579/581(99%), Gaps = 0/581(0%)], [GenBank FJ790854; Identities = 562/563(99%), Gaps = 0/563(0%)].
The RAxML and Bayesian inference analysis showed the similar tree topologies. The tree with the highest log likelihood (-3331.95) is shown in Fig. 2. The new strains from the Niagara Region, Southern Ontario, Canada were grouped with E. vitis specimens from USA and formed a distinct clade with high bootstrap support values (ML/BP=95/1.00).
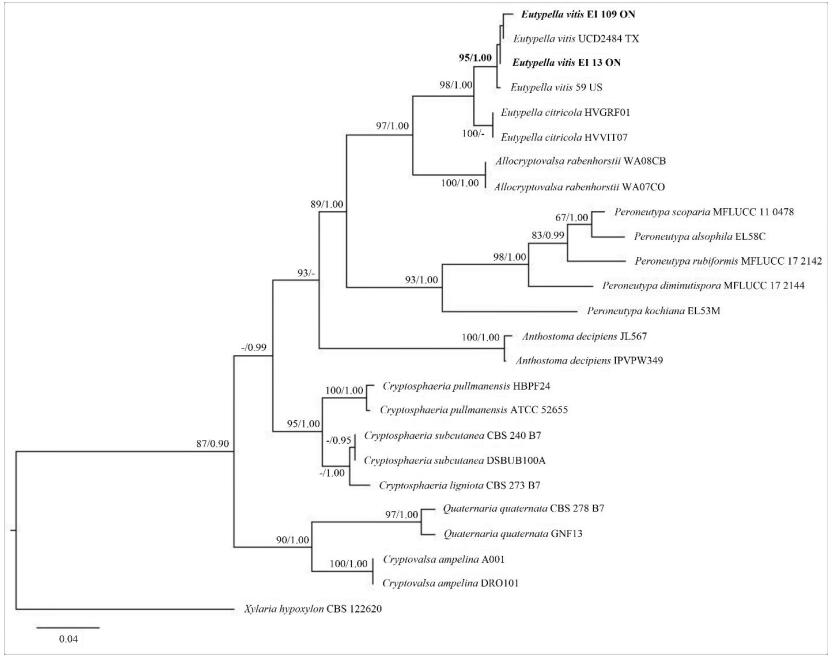
Figure 2. Phylogram generated from the best scoring RAxML tree based on ITS sequence data. Bootstrap support values for ML ≥ 60%, and Bayesian posterior probabilities (BP) ≥ 0.90 are given as ML/BP above or below nodes. Xylaria hypoxylon (CBS 122620) is used as outgroup. The specimens found in this study are in black bold.
Morphological characterization and Taxonomy
-
Eutypa dieback in Canada on Vitis spp. have been previously detected in vineyards surveyed in Okanagan Valley, Vancouver and Gulf Islands and Lower Mainland during the 2011 growing season; the causal agents were identified as E. lata, Eutypa flavovirens and Eutypa laevata (O'Gorman et al. 2013). E. vitis was not found in Canada, however, this species was isolated from diseased grapevines (Vitis labrusca and V. vinifera) with the Eutypa dieback symptoms in several states of USA (Farr et al. 1989, Jordan & Schilder 2005, Vasilyeva & Stephenson 2006, Úrbez-Torres et al. 2009, Jayawadena et al. 2018). This study firstly reports E. vitis on multiply tree hosts (Fagus grandifolia, Fraxinus pennsylvanica and Syringa reticulata) in the Niagara region of Southern Ontario, Canada. The pathogen species identity was confirmed by both morphological identification and phylogenetic analyses of ITS gene region.
According to many researchers, there are two main morphotypes within E. vitis: the first one has shorter ascospores, up to 10.5 µm long (Rappaz 1987), while the second one is characterized by larger ascospores with an average length of more than 10 µm, sometimes up to 14 µm long (Ellis & Everhart 1892, Glawe & Jacobs 1987, Vasilyeva & Stephenson 2006). Our isolates from Canada represent the morphotype with the shorter ascospores (8.5–10 µm). The low virulence of E. vitis in the USA was confirmed based on the results of the pathogenicity tests controlled in the laboratory over a 28-day period (Úrbez-Torres et al. 2009). It is known E. lata slowly colonizes wood while the fungus appears to be a highly virulent pathogen on grapevines (Moller & Kasimatis 1981). Therefore, we can assume that a pathogenicity of diatrypaceous fungi (including E. vitis) has been still underestimated. That can be a critical factor in the field conditions when trees are more likely to be stressed. More surveys are needed to find out if E. vitis has infected grapevines grown in the Niagara region of Southern Ontario. The ability of E. vitis to infect several alternate hosts can contribute to faster and wider spread of the fungus across vineyards in Canada.
- Copyright: © 2021 by the author(s). This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
| E Ilyukhin, T Bulgakov, S Markovskaja. 2021. First record of Eutypella vitis causing branch dieback on new host trees in Canada. Studies in Fungi 6(1):71−77 doi: 10.5943/sif/6/1/3 |












