-
Plant architecture is a critical factor for improving crop yield and reducing labor costs[1]. The selection of compact phenotypes has been a game changer for food security in many crop species[2,3]. Cucurbit crops such as cucumber, melon, watermelon and pumpkin provide plenty of fruits and vegetables for people worldwide. The long and trailing vines of cucurbit plants are an undomesticated characteristic which prevent dense crop planting and labor cost savings. Hence, optimization of cucurbit plant architecture is a key factor for improving crop yield and higher-efficiency agriculture. Recently, we identified a natural regulatory allele, a cucurbit-conserved cis-regulatory element 'B- region' in the 5' UTR of a YABBY1 transcription factor, controls stem length through regulating the translation of YABBY1 in a dose-dependent manner in cucurbit plants[4]. The research established a directed artificial evolution strategy to customize stem length in cucurbit crops, thereby enhancing yield and reducing labor costs. Exploiting artificial quantitative trait loci in the dominant B-region will make it feasible to rapidly domesticate long and trailing vines in cucurbit crops. This work demonstrates the use of rare variants in regulatory elements for crop improvement and further enriches the functional diversity of YABBY family genes, while also reflecting the important application value of YABBY family genes in crop breeding.
Plant hormones usually play an important role in regulating stem length[5]. We showed that the reduced stem of the bushy pumpkin (CmoM-4) was insensitive to exogenous gibberellins (GAs) treatment and inferred that CmoYABBY1 locates downstream of the GAs pathway[4]. To further explore the potential interaction of GA signaling and CmoYABBY1, here we investigated the GAs content and gene expression levels of GAs pathway between the bushy CmoM-4 and WT.
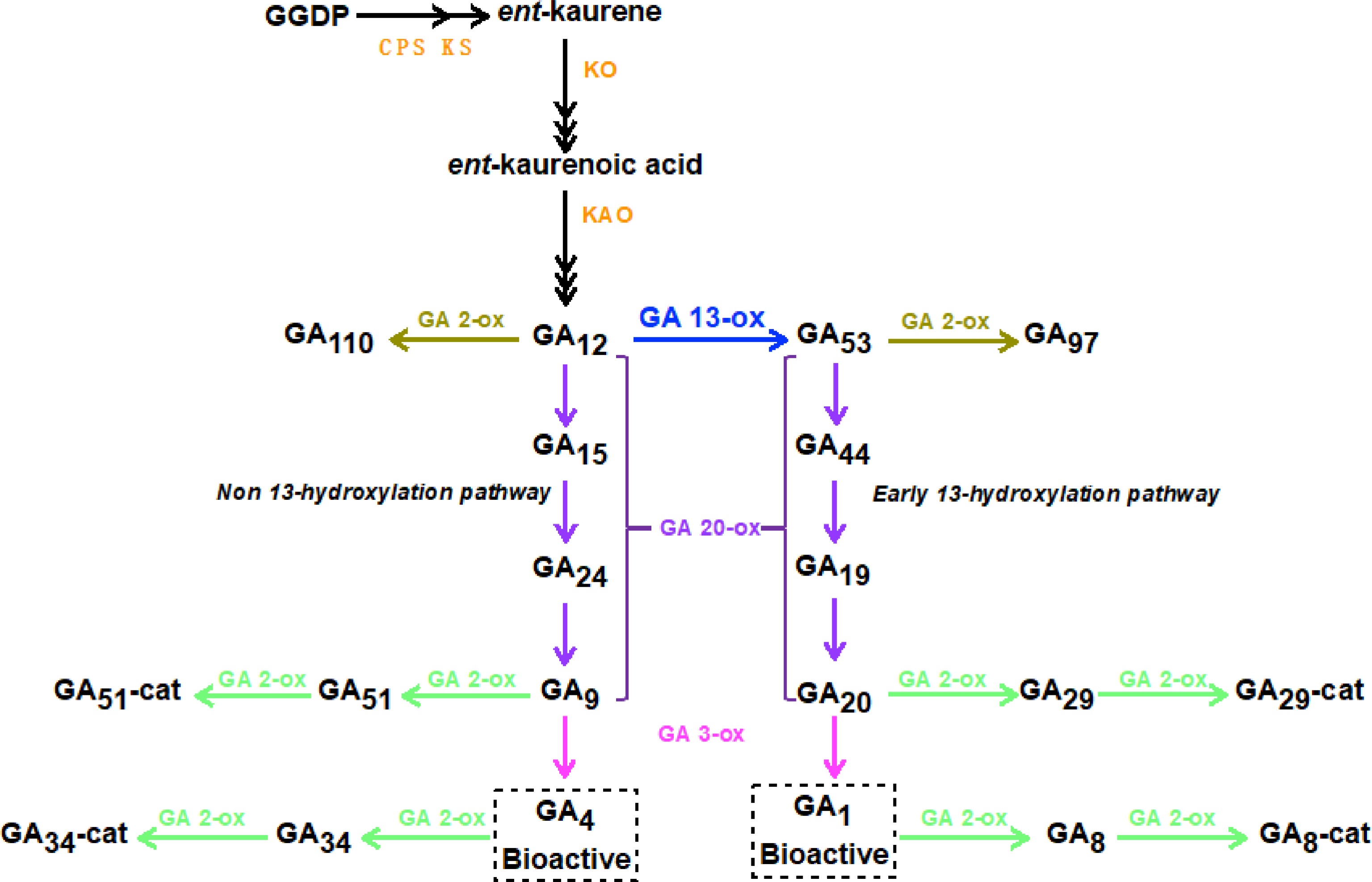
In higher plants, the synthesis of GA is mainly divided into three stages (Fig. 1)[6]. In the plastid, the GA precursor geranylgeranyl pyrophosphate (GGPP) is catalyzed by endo-Cuba pyrophosphate synthase (CPS) and endo-kaurene synthase (KS) to form ent-kaurene. In the endoplasmic reticulum, ent-kaurene is catalyzed by ent-kaurene oxidase (KO) and ent-kaurenoic acid oxidase (KAO) to process into GA12. And in the cytoplasm, GA12 is finally converted into GA1 and GA4 with high biological activity by GA3 oxidase (GA3ox) and GA20 oxidase (GA20ox), respectively. So, we quantified levels of biologically active GAs and bio-inactive GAs in stem tip of CmoM-4 and WT plants using UPLC-MS/MS. As shown in Fig. 2a, for the bioactive GAs (GA1, GA3, GA4, and GA7), we did not detect GA1 and GA3 in any genotype, and the levels of GA7 were not significantly different between two genotypes. While the levels of GA4 were significantly higher in the bushy mutant CmoM-4 than WT. For the bio-inactive GAs, we found that the levels of GA12 were significantly higher in the bushy mutant CmoM-4 than WT plants, and no significant differences in levels of other bio-inactive GAs were detected.
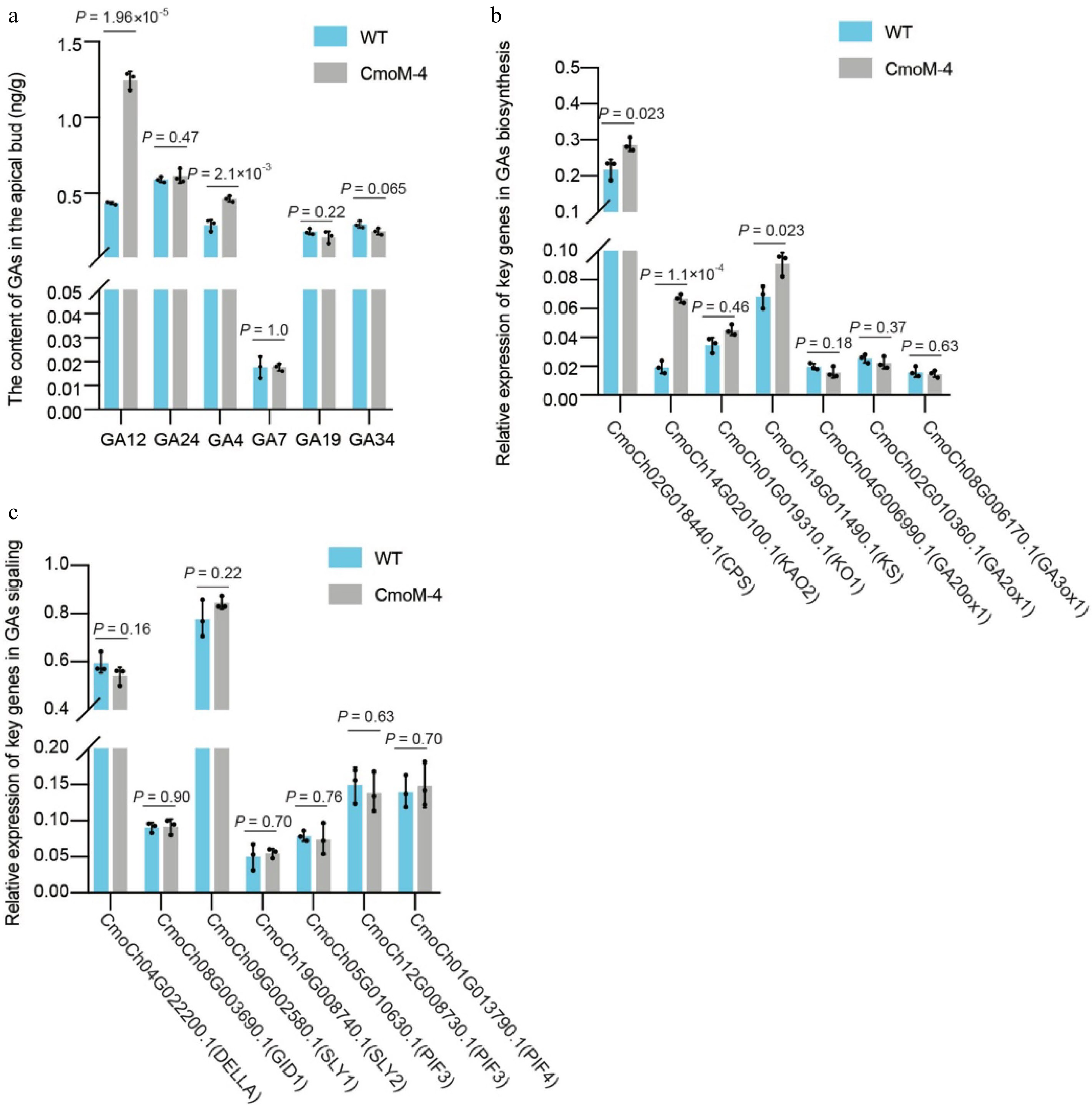
Figure 2.
The content of GAs in apical buds, the relative mRNA expression of key genes in GAs biosynthesis, the relative mRNA expression of key genes in GAs signaling between WT and CmoM-4
We further perform quantitative real time PCR (qPCR) to detect the expression levels of genes related to GA biosynthesis and signaling in stem tips of CmoM-4 and WT plants. We showed that the expression of CPS gene, KS gene, KO1 gene and KAO gene were significantly up-regulated in CmoM-4 compared to WT, which is consistent with the increased levels of GA12 and GA4 in CmoM-4 mutant. While the expression levels of GA2ox-1, GA3ox-1, GA20x-1 did not show difference between two genotypes (Fig. 2b). Furthermore, we detected the expression of GA signaling pathway genes and also the GA target genes. The results showed that the GID1 gene, DELLA gene, SLY gene, as well as GA target gene PIF3 and PIF4 did not show significant differences in the expression levels between CmoM- 4 and WT (Fig. 2c). These results suggested that the increased translation level of YABBY1 protein in CmoM-4 may disturb the downstream signaling of GA in an unknown regulatory mechanism, which in turn enhanced GA biosynthesis in a feedback regulation manner.
How does YABBY1 translation affect the intercalary growth of cucurbit crops? It is reported that auxin is transported towards the SAM from lateral organs to maintain SAM organization in Arabidopsis[7], and we inferred that the bushy phenotype with smaller SAM can be attributed to the increased translation of CmoYABBY1 which enhances the auxin flow from the leaf to the SAM[4]. The regulatory mechanism of CmoYABBY1 and its relationship with hormones are still unknown and complex, it would be important to further understand the regulatory mechanism in future work.
HTML
-
The authors declare that they have no conflict of interest.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
| Wang S, Yang X. 2023. Gibberellins-independent stem length regulation by YABBY1 in cucurbit crops. Vegetable Research 3:11 doi: 10.48130/VR-2023-0011 |